Part:BBa_K3610030
BAK1/YFP
This part can be used for expressing the plant pattern recognition receptor BRI1-associated receptro kinase (BAK1) from Arabidopsis thaliana in S. cerevisiae. This part contains the full length receptor BAK1, including the sequence for the signal peptide native to A. thaliana (BAK+). To make expression of BAK1 visible and to test observe the localization in the cell, the BAK1 coding region has been fused to a yellow fluorescent protein by a 15 amino-acid long linker.
Usage and Biology
BAK1 is a cell surface receptor protein with an intracellular kinase domain and an extracellular ligand binding domain. The receptor is necessary for many functions in the plant like brassinosteroid signalling and it is also a critical player in plant immunity, as BAK1 interacts with many important cell surface receptors which perceive pathogen-associated molecular patterns (PAMPs). One example of these PAMPs is the 22-amino-acid peptide flg22 from flagellin which is recognized by the leucine-rich repeat receptor kinases flagellin-sensitive 2 (FLS2). Upon recognizing the flg22 peptide, FLS2 interacts with BAK1. This interaction drives the immune response of the plant.
In our project, we used this part to test the expression of the whole length receptor in S. cerevisiae, as well as to observe the localization of the protein within the cell. It is important to note that the protein coding domain of this part has its original signal sequence from A. thaliana.
Characterization
Expression of BAK1/YFP in S. cerevisiae
In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed E. coli (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same E. coli strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different strength to get more quantitative data. Finally, we got the construct in a plasmid with a truncated version of the ADH1 promoter from S. cerevisiae. For termination, this part has the terminator sequence of the enolase 2 protein from S. cerevisiae. The plasmid also contained the TRP1 gene, which encodes phosphoribosylanthranilate isomerase, an enzyme that catalyzes the third step in tryptophan biosynthesis. This enabled us to use the same plasmid for expression in S. cerevisiae. We prepared a medium containing YNB and free amino acids, without tryptophan. S. cerevisiae cells (AP4) were transfected with the plasmid and then plated on the selective medium.
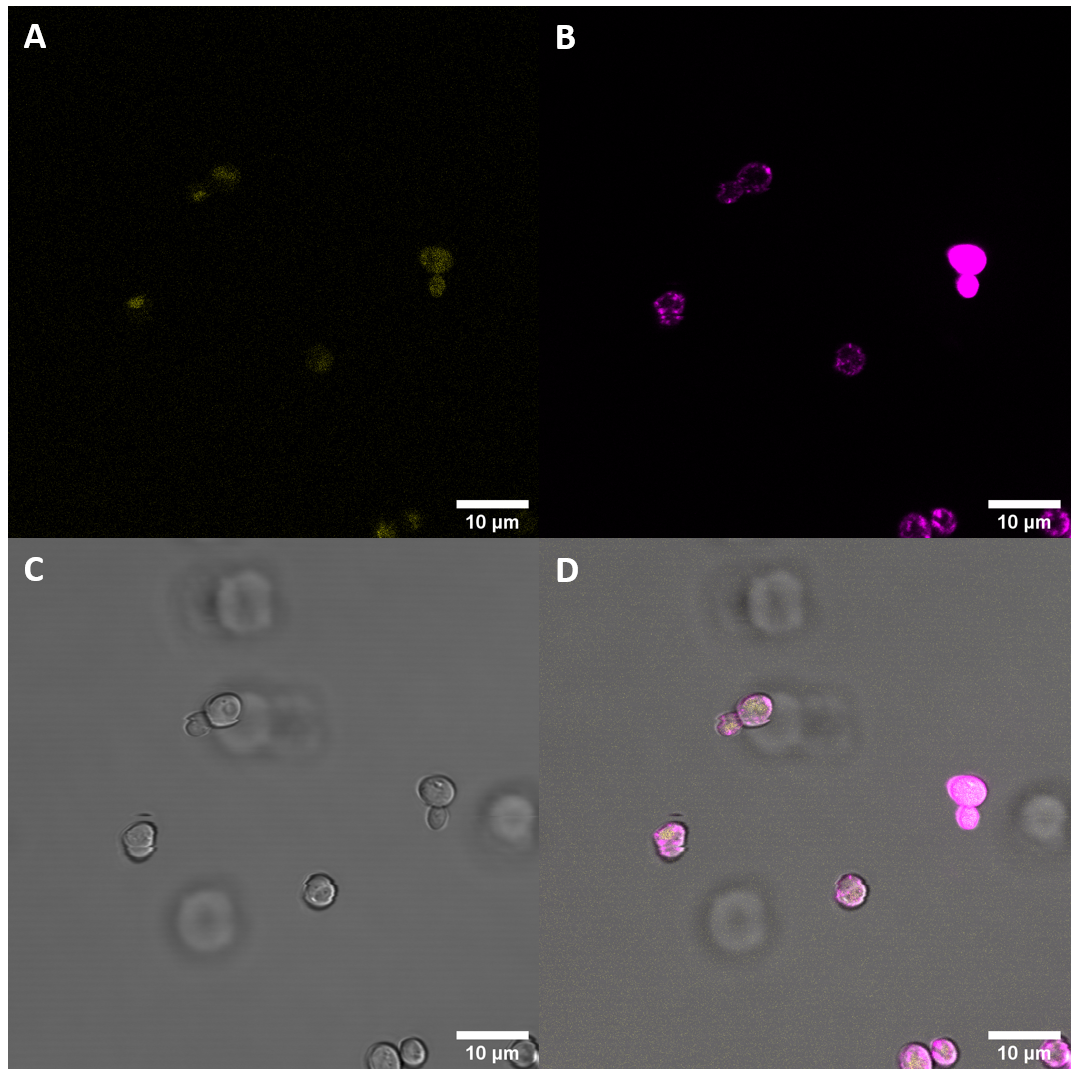
Confocal Fluorescence Microcopy
After successful transfection of S. cerevisiae cells with the plasmids we exmined the cell cultures with fluorescence microscopy.
Observed fluorescence with fluorescence microscopy was weak and did not appear to be localized at the plasma membrane (PM). As localization at the cell membrane was something we were particularily interested in, we repeated the confocal microscopy step with an additional membrane stain. The cell membrane was stained with fm4-64, which fluoresces strongly after binding to the cell membrane ((λEX = 515nm and λEM = 640nm). The binding of the dye is happening rapidly and it is also reversible. If the time spent between staining and imaging is too long, then the dye will be taken up by the organism and stored in the vacuole. Imaging with a confocal microscope for YFP and the fm4-64 stain shows the spatial overlap of the red fluorescence of the stain and the yellow fluorescence of the protein fused to the receptors.
Fluorescence Microscopy suggests that there is some expression of the BAK+ construct. It fails, however, to get localized at the cell periphery. Nevertheless, expression of the full length receptor in S. cerevisiae can already be seen as a considerable success.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 1000COMPATIBLE WITH RFC[1000]
| None |