Part:BBa_K3260027
ZE Leucine Zipper
Leucine Zipper, constitutes one part of the heterodimeric pair with ZR Leucine Zipper, BBa_K3260029. This part and its dimeric partner were based off of a leucine pair similar to the B-ZIP system in humans (originally designed by Zhang et al [2]), but was further modified to enhance protein affinity by Moll et al in 2005 [1]. This part is taken from Tirrell’s paper, but was separated from its endogenous linker.
[1] Moll, Jonathan R., and Sergei B. Ruvinov . “Designed Heterodimerizing Leucine Zippers with a Range of PIs and Stabilities up to 10-15 M.” Protein Science , Cold Spring Harbor Laboratory Press, 2001.
[2] Park, Won Min. Self-Assembly of Protein-Based Suprastructures . Georgia Institute of Technology School of Chemical & Bimolecular Engineering , May 2015.
[3] Zhang, Kechun, et al. Artificial Polypeptide Scaffold for Protein Immobilization . American Chemical Society , 2005.
Usage and Biology:
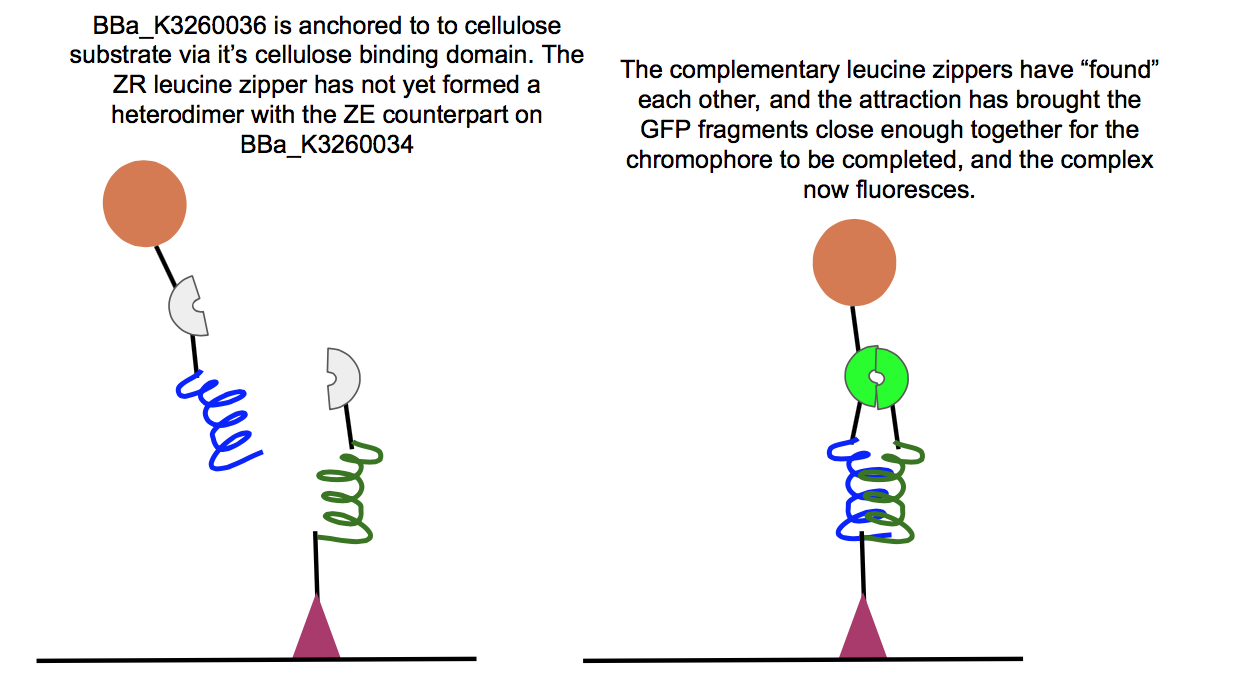
This part was used in the composite BBa_K3260034, in conjunction with the BBa_K3260036 composite. Below are the results for experiments regarding the interaction between the two composites.


Above is a test of the functionality of two of our new fusion proteins, BBa_K3260036, and BBa_K3260034. If these two parts assemble correctly on the cellulose surface, there should be fluorescence, which is what we observed here. This means that the leucine zipper components of each protein aligned and held together, allowing the two halves of GFP to get close enough together to complete their chromophore and fluoresce. Because we see fluorescence (highlighted in the color swatch of each sample), the success of the leucine zippers and therefore the functionality of the last two functional domains can be inferred as well, the double cellulose binding domain anchoring the whole complex to the cellulose surface, as well as the hydrophobic domain sticking up from the N-terminal region of BBa_K3260034, and therefore the assembled protein complex. The successful assembly of these two biobricks not only shows the applicability of these constructs for paper-based microfluidics, but it also shows the potential of split-GFP reporting for fusion proteins in functional applications.
</p>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
