Part:BBa_K3128018
OmpX-WT protein fused with T25 subpart of Bordetella Pertussis AC under constitutive promoter
Contents
Sequence and features
This biobrick is composed of the external membrane Outer Membrane Protein X Wild Type (OmpX-WT)
protein fused at its N-terminal end to a 54 aa Gly-Gly-Ser Linker (GGS)
followed by the T25 sub-part of the Bordetella Pertussis adenylate cyclase (AC) .
This biobrick is under a constitutive promoter J23111,
thereby OmpX-WT fused to T25 protein is constitutively expressed leading to a large amount of recombinant protein in the outer membrane at any time.
This BioBrick is intended to operate with BBa_K3128017.
These two biobricks are the negative condition of the mBACTH -free sub-parts condition-.
OmpX proteins are fused to the adenylate cyclase sub-parts at their N-terminal ends. The fusion protein move freely in the bacterial outer membrane,
but they are not forced to get closer by any mean.
The reconstitution of the adenylate cyclase in this condition is only due to random occurrence between both parts.
The signal measured is considered as background noise.
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1471
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
In 1989, Fields and Song demonstrated a new genetic system allowing the detection of protein-protein interaction (1). At first, it was performed in Saccharomyces cerevisiae
yeast and it was named the yeast two-hybrid assay (Y2H). In 1998, Ladant and al. described the system in bacteria (2). Nowadays, this biological technique is mostly
used to show and characterize the physical interaction between two cytosolic proteins or internal membrane proteins in vivo (3).
Bacterial Adenylate Cyclase Two-Hybrid (BACTH)
The principle lies on the interaction-mediated reconstitution of a signalling cascade in Escherichia coli. The messenger molecule involved in this cascade is the
cyclic adenosine monophosphate (cAMP) produced by the adenylate cyclase. Adenylate cyclase is an enzyme catalysing the cAMP production from ATP. It
physiologically participates to the cellular transmission.
This system involves the Bordetella pertussis adenylate cyclase. You might know this bacterium; it is the responsible agent for the pertussis disease.
It adenylates cyclase catalytic domain has the particularity to be splittable in two distinct parts: T18 and T25 sub-parts, unable to work unless they
reassociate. Each sub-part of the enzyme is fused with a protein of interest, either the bait or the prey protein chose beforehand by the experimentator.
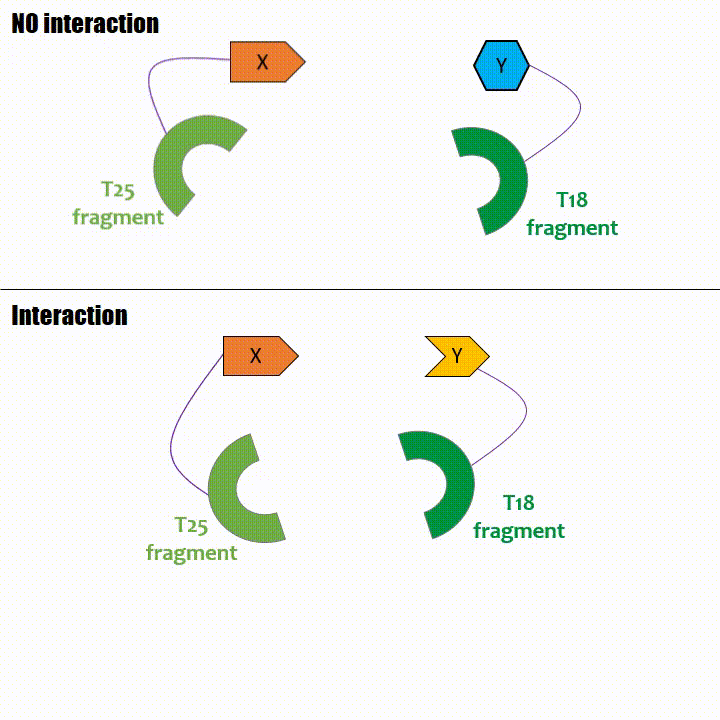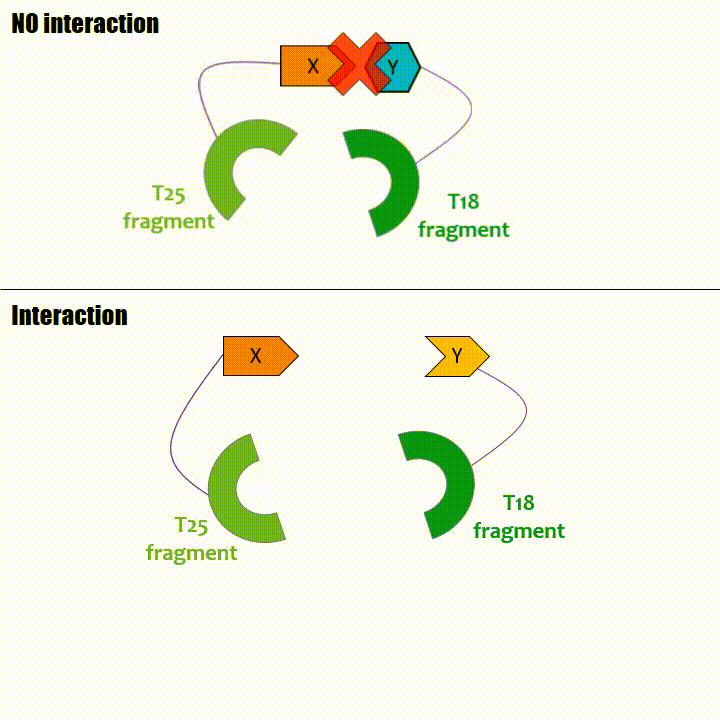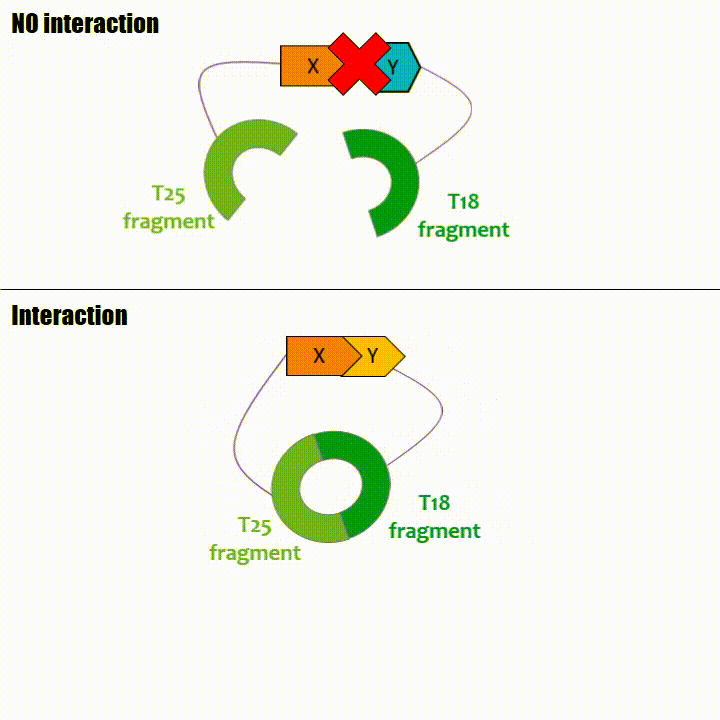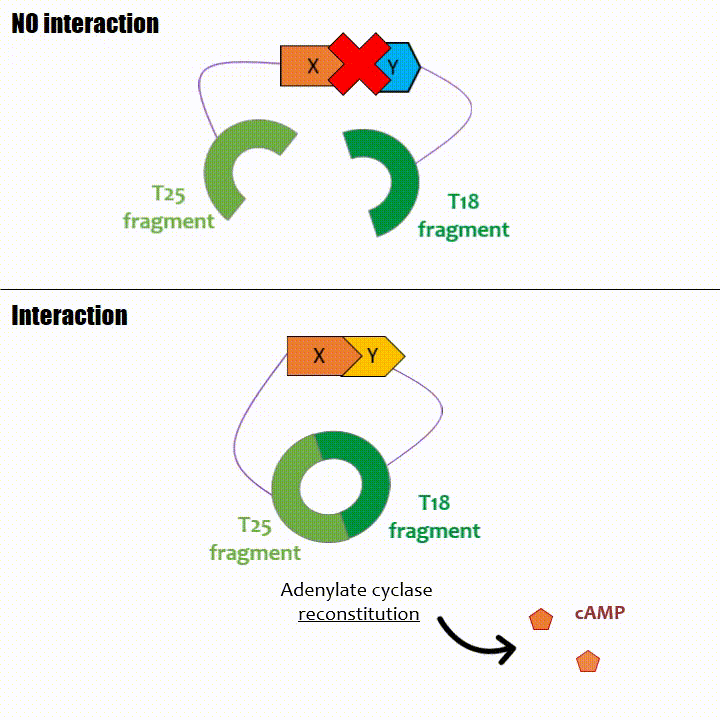
If two proteins interact, then T18 and T25 are bring together and reconstitute a functional adenylate cyclase enzyme thus enabling cAMP production. Using
cya- bacteria – strain for whom the adenylate gene is deleted, involving an absence of this endogenous enzyme – a BACTH could be done with the creation of two
fusion proteins : the first one, fused at its N or C terminal intracellular end with the T18 sub-part; the second one fused with the T25 sub-part.
The interaction of these proteins of interest will lead to the adenylate cyclase reconstitution, thus initiating cAMP production. The cAMP produced will act as a
messenger by fixing itself to the transcriptional activator CAP, cAMP form the CAP-cAMP complex, controlling the expression of the lactose promoter by initiating transcription of the following gene.
This promoter is placed upstream the chosen reporter gene.
NeuroDrop Project - Outer-Membrane BACTH (mBACTH)

[http://2015.igem.org/Team:TU_Eindhoven Eindhoven-2015] iGEM project’s aim was to develop a “universal membrane sensor platform for biosensors”.
This year, Team Grenoble-Alpes is designing a new tears biosensor system based on [http://2015.igem.org/Team:TU_Eindhoven Eindhoven-2015]’s project.
Both projects have a common base, the same receptors are used at the external surface of bacteria : Clickable Outer Membrane Protein X(COMP).
OmpX is an outer membrane protein with the C- and N-termini in the intracellular domain. To be able to use OmpX as a scaffold, a unnatural amino acid needs to be introduced.
This can be done by implementing the amber stop codon TAG in one of the loops of OmpX via a mutation. With a specific tRNA an azide-functionalized amino acid can be built in,
which can be used for the SPAAC click chemistry reaction using DIBO functionalized groups, this modified protein is called COMP.
The complex aptamer fixed to a COMP is then named a COMB for Clickable Outer Membrane Biosensor.
The Grenoble-Alpes team aims to develop an Outer membrane Bacterial Adenylate Cyclase Two Hybrid (mBACTH).
In this case, the two adenylate cyclase sub-parts are fused to the N-terminal ends of COMPs with a Gly-Gly-Ser Linker (GGS) of 54 amino acids - in order to ensure a sufficient flexibility -.
When COMBs catch the extracellular target, they get closer, thus allowing the reconstitution of a functional adenylate cyclase due to the physical proximity of
the two sub-parts.
The enzyme is operational again and produce a high quantity of cAMP (around 17,000 mmol of cAMP formed per mg of adenylate cyclase per minute),
the molecule responsible for the signal transduction in the bacteria.

cAMP molecules diffuse to the cytoplasm of the bacterium and interact with catabolite activator proteins (CAP) in a ratio 1 to 1.
Yhen two cAMP-CAP complexes are needed to activate the expression of the gene under the control of the lactose promoter.
Because of the high quantity of cAMP diffusing in the cytoplasm of the bacterium (2), the reporter gene is continously activated as long as cAMP is produced.

The high enzymatic activity (1) of Bordetella pertussis Adenylate Cyclase generates a high production of cAMP in presence of ATP in the bacterium
thus activating the signalling cascade with the CAP-cAMP dependant promoter.
Hence this system is promising because it might have a great sensitivity and may drive a great signal amplification for a low amount of melocules to be detected.
References
(1) Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature [Internet]. 1989
(2) Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. PNAS [Internet]. 1998
(3) Karimova G, Gauliard E, Davi M, P.Ouellette S, Ladant D. Protein–Protein Interaction: Bacterial Two-Hybrid. 2017
//cds/membrane/extracellular
//cds/membrane/receptor
//classic/signalling/sender
//rbs/prokaryote/constitutive
| sender |
