Part:BBa_J23100
constitutive promoter family member . For video explanation on this promoter family and its use, see [http://www.synbiotrails.org/#!/trail?id=org.clothocad.trails.youtube.TranscriptionModels this].
Variant RFP (au) J23112 1 J23103 17 J23113 21 J23109 106 J23117 162 J23114 256 J23115 387 J23116 396 J23105 623 J23110 844 J23107 908 J23106 1185 J23108 1303 J23118 1429 J23111 1487 J23101 1791 J23104 1831 J23102 2179 J23100 2547 |
Constitutive promoter family
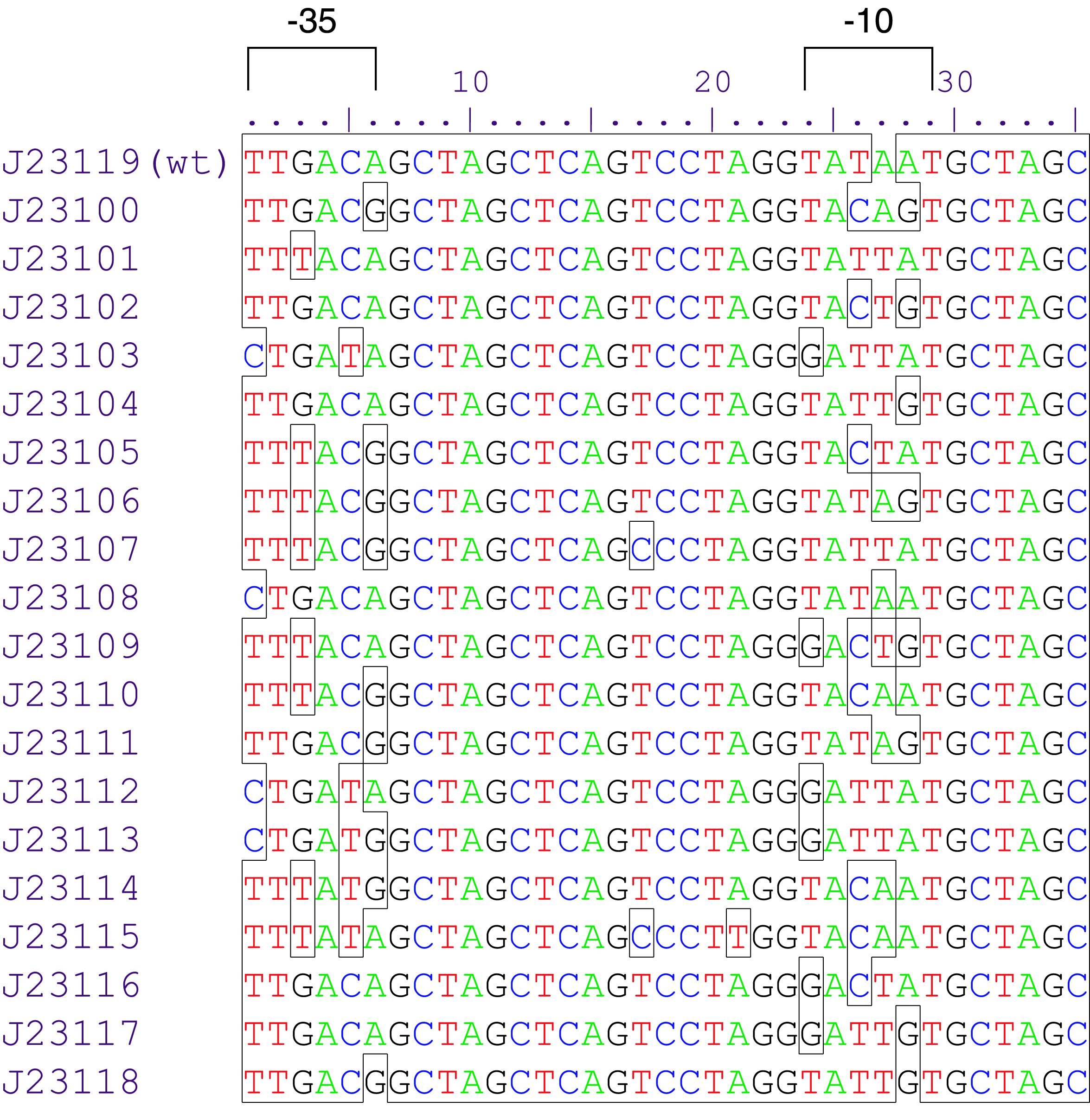
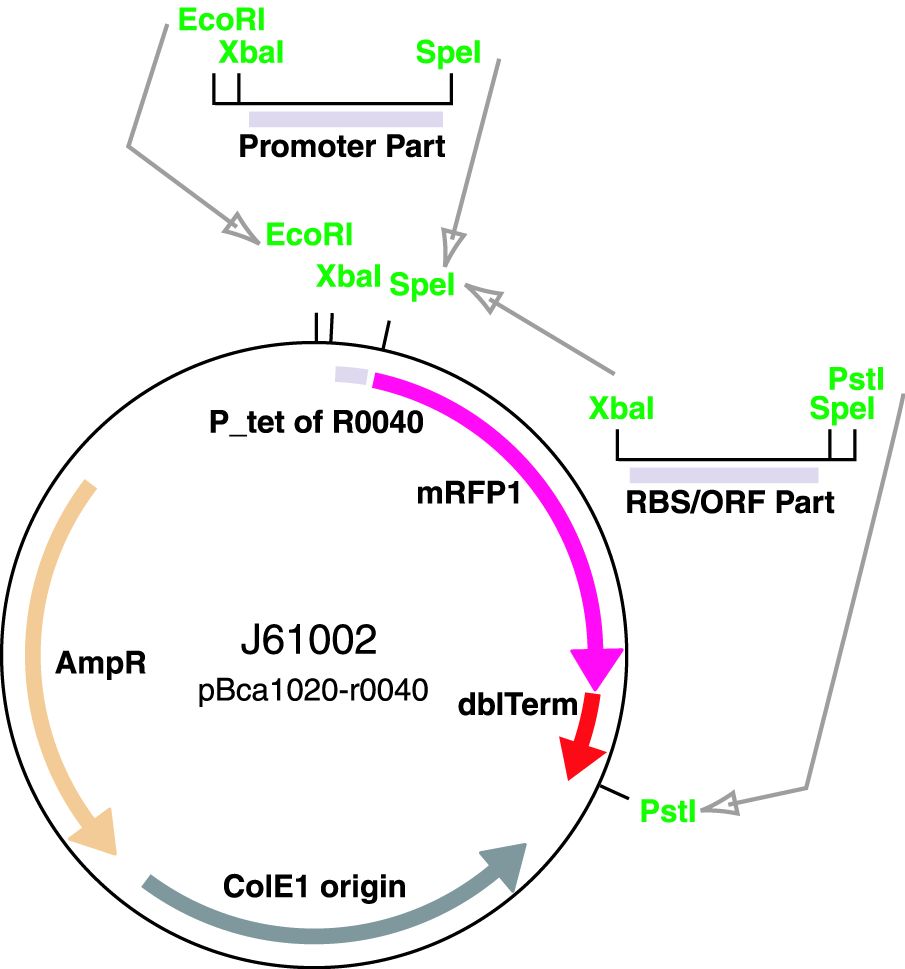
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part BBa_J61002 for details on their use.
These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. JCAraw
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
Relative promoter strength estimates (see [http://2009.igem.org/Team:Groningen/Promoters this page] from Groningen 2009):
| Reference | Strength |
|---|---|
| BBa_J23109 | 67 |
| BBa_J23106 | 2 |
USTC_2009's MEAUREMENT
[K176011]
Usage and Biology
The Yale iGEM team has assembled this promoter to citrine (an improved version of YFP, with excitation peak at 514nm and emission peak at 527nm) and a T7-terminator to quantify the level of expression in E. coli and in non-model organism hosts. This construct is available as the biobrick K185000
This construct has been successfully cloned into E. coli using the broad-host range vector pKT230, a RSF1010 derived plasmid, as well as using the pPZP200 plasmid which can be transformed into agrobacterium and rhizobium. Leaky expression of citrine was observed.
Yale 2015's Characterization
Measured strength
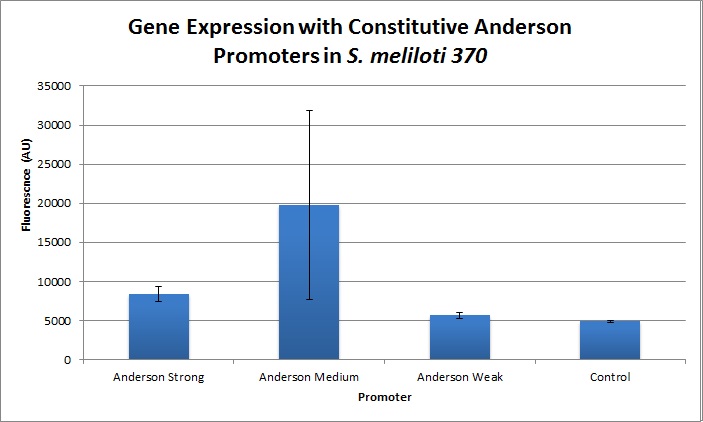
The Yale iGEM team found fluorescence levels nine times above the baseline level in E. coli cells transformed with the promoter-citrine construct. We also found this promoter to be effective in driving gene expression in S. meliloti, with fluorescence levels of over two times that of the baseline observed.
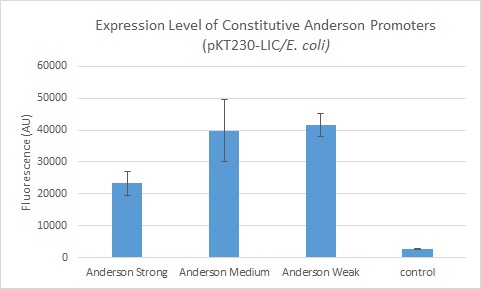
We further compared expression level of this promoter to two of the other promoters in the Anderson collection. The Anderson lab found relatively expression levels of 1, 0.33, and 0.10 respectively when comparing the promoters J23100 (Anderson Strong), J23110 (Anderson Medium) and J23114 (Anderson Weak). We found the relative expressions to be 1, 1.71 and 1.78 when we transformed our constructs into E.coli: K1856004 (Anderson Strong-Citrine), K1856003 (Anderson Medium-Citrine), K1856002 (Anderson Weak-Citrine). This highlights that even as all three Anderson promoters are effective in driving gene expression in E. coli, the relative expression level may vary depending on the gene fused to the promoter and on the E. coli strain used. The Yale iGEM team's readings were obtained in E. coli DH10B.
Comparing expression levels in S. meliloti (a rhizobium strain), the relative expression values were 1, 2.36, and no significant expression for the Anderson Strong, Anderson Medium, and Anderson Weak promoters respectively.
Team Warsaw 2010's measurement
Absolute promoter strength: 13,1pg RNA/minute/ug substrate DNA. It equals 2,80 microPoPSSheffield 2016's Characterisation
Measured strength
Sheffield 2016 has improved the characterisation of both BBa_J23100 and BBa_J23106. These parts are a strong and medium promoter respectively, that we have used to design our iron detecting device. We have experimentally validated through fluorimetry that there is indeed a significant difference between expression levels of GFP coupled to the strong and medium promoters. Comparative analysis of promoter strengths can be directly interpreted from the data we obtained. This data can be found both on the original part experience pages of BBa_J23100 and BBa_J23106, as well as at 2016.igem.org/Team:Sheffield/Parts. Our team has also improved the function of the BBa_J23100 part, by adding an RBS site downstream of the promoter (BBa_K2016005). BBa_J23100 was originally submitted to the Registry by Berkeley 2006.

Fluorescence of JC28 mutants or W3110 wild types transformed with RyhB-GFP constructs under the control of medium (MedGFP) or strong promoters (StrGFP).
//direction/forward
//promoter/anderson
//regulation/constitutive
//rnap/prokaryote/ecoli/sigma70
| efficiency | 8.5326 molecule s^(−1 ) (per cell) |
| n/a | constitutive promoter family member |
| negative_regulators | |
| positive_regulators |

 1 Registry Star
1 Registry Star