Part:BBa_K1732000
J23100-PelB-GlucHCO-B0015
J23100 Promoter with a RBS (BBa_S04423), PelB Leader sequence, Gaussia princeps luciferase, and a B0015 terminator. Allows Gaussia princeps luciferase to be transcribed and translated using a strong promoter. Luminescence can be expressed by reacting the luciferase product with coelenterazine.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 546
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 115
- 1000COMPATIBLE WITH RFC[1000]
Gaussia Luciferase (GLuc) is a naturally secreted protein from Gaussia princeps. It is considered the brightest luciferase and its size is around 19kD. This luciferase is stable at high temperatures due to the presence of disulfide bonds in its structure. This stability makes it an ideal reporter gene, specifically for coelenterazine.
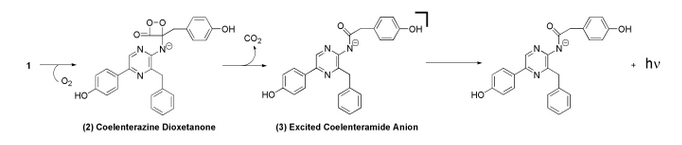
Bioluminescent Mechanism of Coelenterazine
Coelenterazine is found in several aquatic organisms and is a luminescent substrate for many luciferase enzymes. For this study, PelB Gaussia Luciferase interacted with coelenterazine to determine relative light output and kinetics.
The image above is the chemiluminescent mechanism of coelenterazine. The bioluminescent mechanism is similar, except that the excited state molecule in bioluminescent is phenolate anion instead of the amide anion.
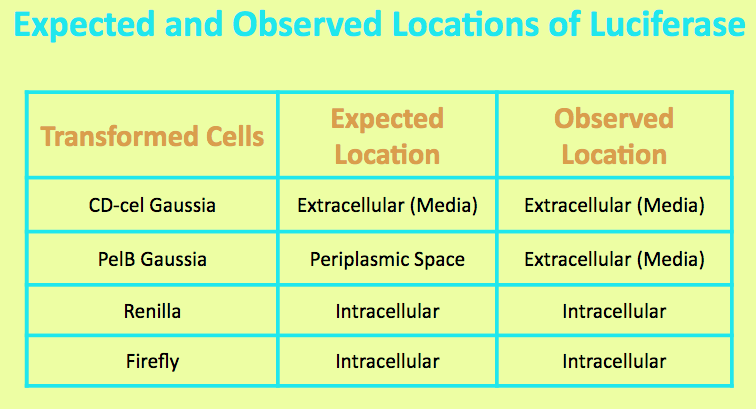
Before running experiments with Gaussia Luciferase, the location of the luciferase in the media was determined by comparing the lightout of overnight grown culture of PelB Gaussia cells with those of the pellet and the supernatant. The PelB Gaussia culture was spun down, and the pellet represented intracellular localization while the supernatant represented extracellular localization.
//proteindomain/localization
//rbs/prokaryote/constitutive/constitutive
| None |