Part:BBa_K1321340
Double CBD (dCBD) with N-terminal linker
Double cellulose binding domain (dCBD) using two cellulose binding domains from Trichoderma reesei cellobiohydrolases, with an N-terminal linker and internal linker sequence between the two domains which is derived from the endogenous cellobiohydrolase linker sequence.
Usage and Biology
This part is based on the double cellulose-binding domain construct (CBDcbh2-linker-CBDcbh1) synthesised and characterised by Linder et al [http://dx.doi.org/10.1074/jbc.271.35.21268 (1)] who found that this double CBD had higher affinity for cellulose than either of the two CBDs on their own. The main difference is that our part contains an additional linker sequence on the N-terminus of the protein.
The two CBDs are from the fungus T. reesei (Hypocrea jecorina) Exocellobiohydrolase (Exoglucanase) I (cbh1), uniprot ID [http://www.uniprot.org/uniprot/P62694 P62694]; and Exocellobiohydrolase (Exoglucanase) II, uniprot ID [http://www.uniprot.org/uniprot/P07987 P07987] (cbh2); with a linker peptide between the two CBDs and at the N-terminus of the protein. Both linkers are the same amino acid sequence and are based on the endogenous linker sequences that exists in cbh1 and cbh2 genes. The linker sequence is PGANPPGTTTTSRPATTTGSSPGP which is the same as used by Linder et al [http://dx.doi.org/10.1074/jbc.271.35.21268 (1)]. The first three amino acids are from the cbh2 endogenous linker, and the rest is from the cbh1 endogenous linker. CBDcbh1 is placed C-terminal to CBDcbh2 because naturally CBDcbh1 is a C-terminal domain and CBDcbh2 is an N-terminal domain. Both CBDs are from the [http://www.cazy.org/CBM1.html CBM family 1]. The precise location of the CBD within the cbh genes was slightly different according to the uniprot annotations and the sequence used by Linder et al [http://dx.doi.org/10.1074/jbc.271.35.21268 (1)]; we chose to use the sequence from the paper since the protein was expressed and characterised successfully.
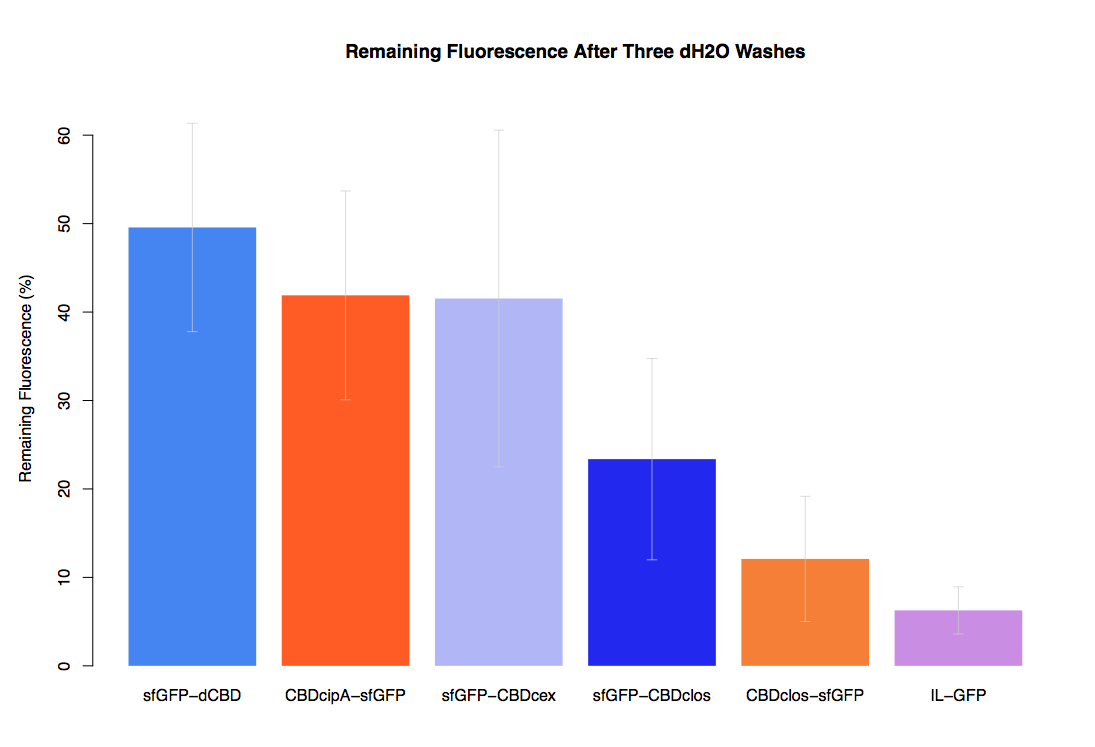
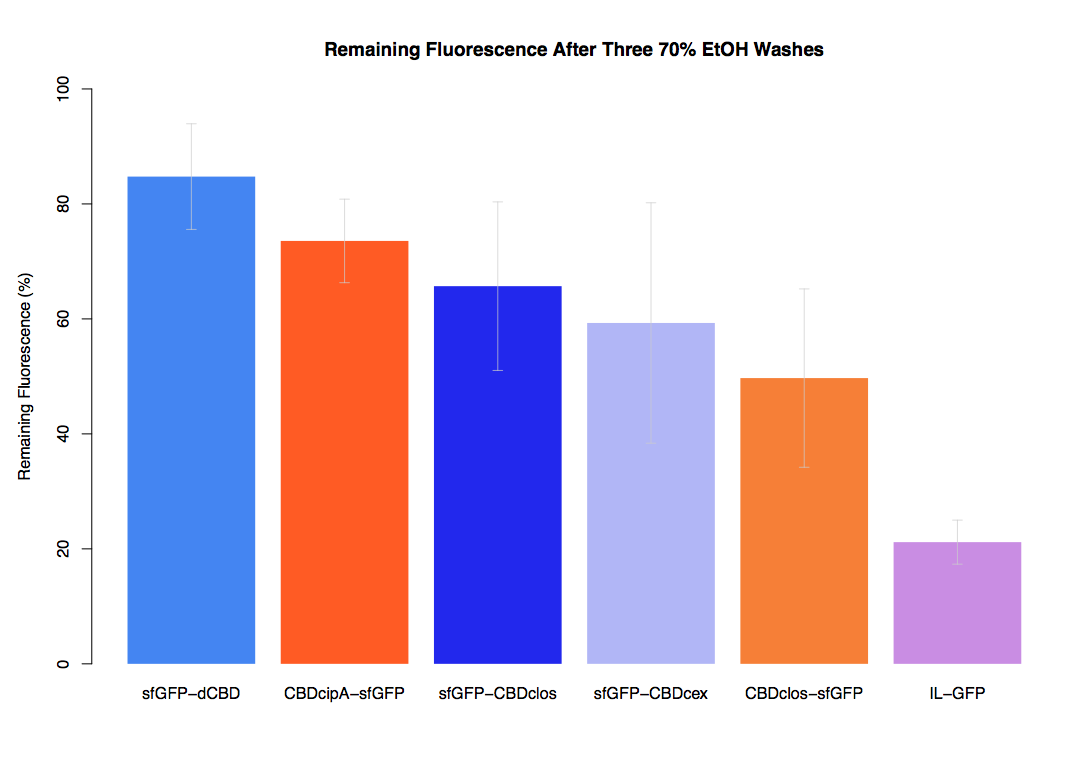
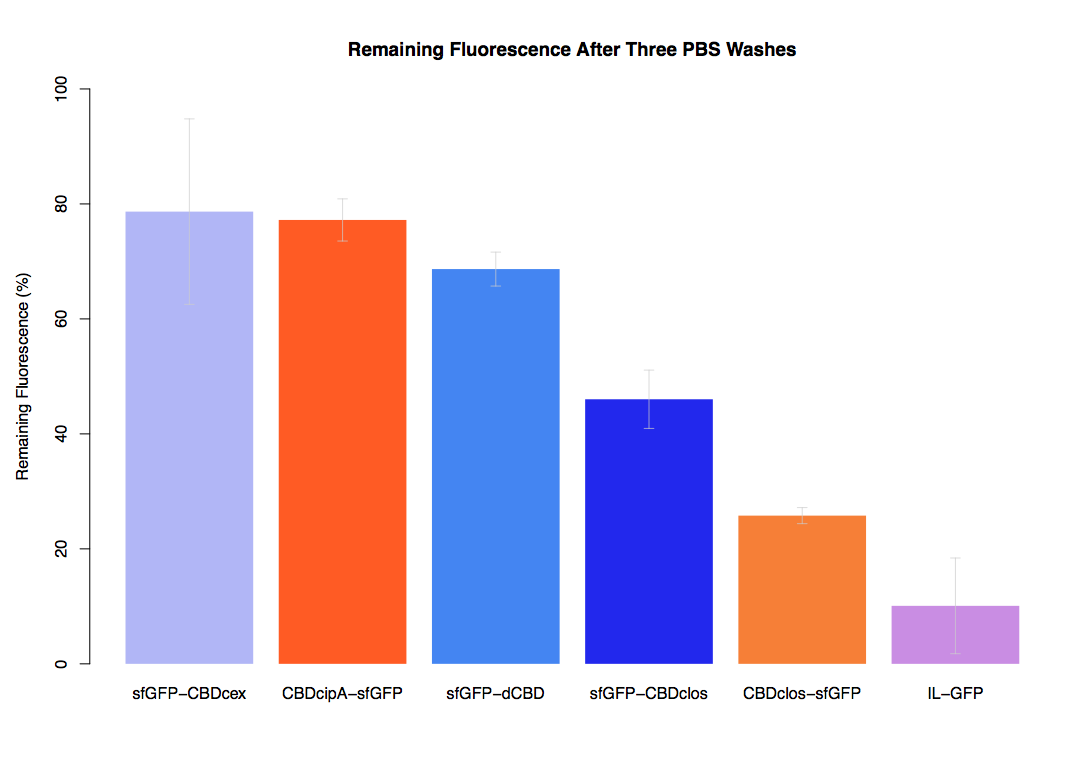
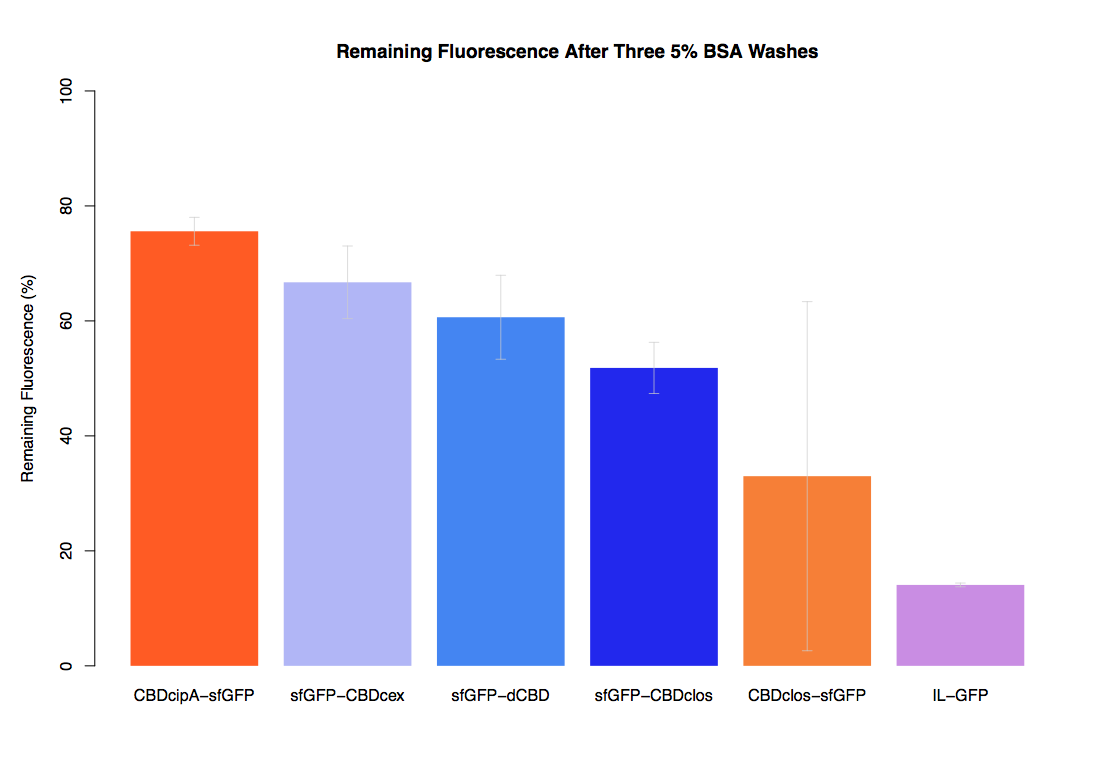
As part of our project we carried out an assay to determine the relative binding ability of CBD-sfGFP fusions to bacterial cellulose.
In our first assay performed to determine the relative strengths of various CBDs’ binding to bacterial cellulose – represented by the percentage fluorescence left from CBDs fused to (sfGFP (RFC25)) bound to bacterial cellulose discs, when subjected to various washes - it was determined that the fusion with dCBD had the greatest binding ability in comparison to four other CBDs fused to sfGFP after three washes with both dH2O and 70% EtOH.
In the same assay, results suggested that dCBD had on average the third greatest ability to bind bacterial cellulose, after three washes with both PBS and 5% BSA.
It was thus decided that dCBD was one of the top three strongest CBDs and was carried forward for use in a metal-binding assay, fusing it to synthetic phytochelatin (insert part number) to show the successful functionalisaton of bacterial cellulose as a metal-removing filter.
Please see below for AutoAnnotator information about the final protein product.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_K1321340 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25: (underlined part encodes the protein) ATGGCCGGC ... CTTACCGGTTAA ORF from nucleotide position 1 to 381 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
References
1. Linder, M.; Salovuori, I.; Ruohonen, L.; Teeri, T.T., 1996. Characterization of a Double Cellulose-binding Domain. SYNERGISTIC HIGH AFFINITY BINDING TO CRYSTALLINE CELLULOSE. Journal of Biological Chemistry, 271(35), pp.21268–21272. Available at: http://www.jbc.org/content/271/35/21268.full
| None |