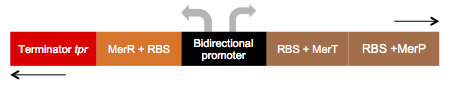
Part:BBa_K1355001
Regulation and transport of mercury ions
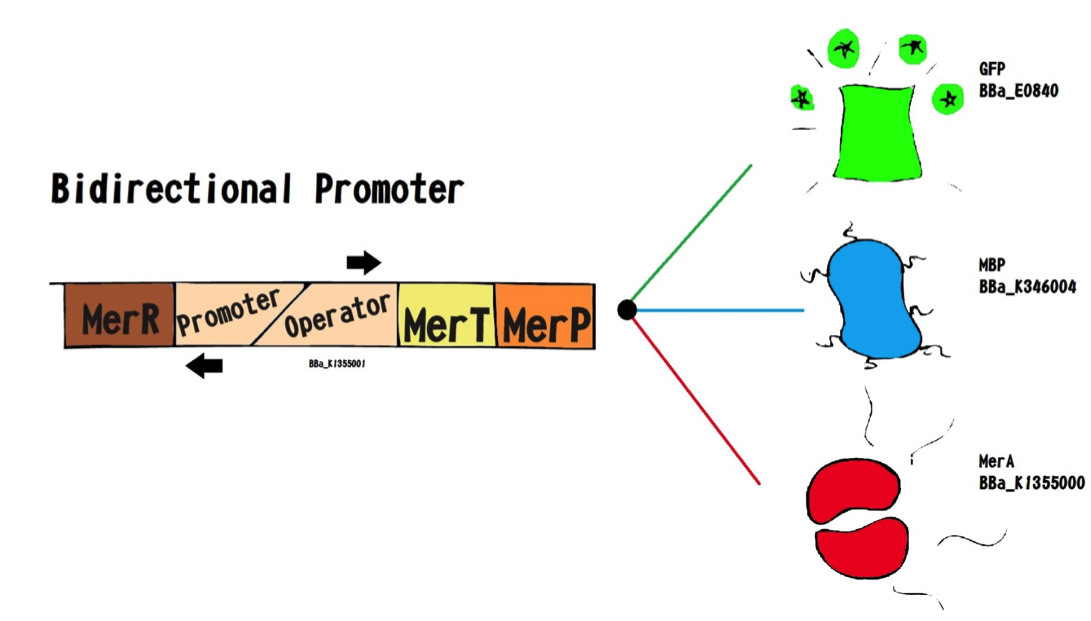
We call this part an “Essential biobrick”. We designed it meaning to be the key piece of many genetic construction related to mercury, eg. Bio-sensor, bio-remediator, bio-accumulator, and among others. This biobrick has bidirectional promoter, having dual functions: A) In reverse: transcription of the MerR regulator protein; and B) In forward: transcription of the MerP and MerT proteins, as represented below:
The operon expression regulation is performed by MerR protein. In the absence of mercury, merR forms a merR-promoter-operator complex, preventing RNA polymerase to recognize the promoter consequently, messengers RNA MerPT will not be transcript. In the presence of Hg 2+ ,MerR protein binds to this element and dissociates from the promoter-operator complex, allowing MerPT expression.
The MerP is a carrier protein that is located in the periplasmic space and has about 91 amino acids. It binds and transports mercury from the inner membrane to the periplasm, the next mercury transport protein is MerT. The MerT has about 116 amino acids and is also a carrier protein it is located in the inner membrane and when binds to mercury transport it from the inner membrane to the cytoplasm.
Essential biobrick (BBa_K1355001) doesn’t have a transcription terminator, because it was made to be attached to others coding regions that permit a new function for bacteria. For this project, we used Escherichia coli DH5-alpha with Essential Biobrick attached to encoding region for Green Fluorescent Protein (BBa_E0840) to build a biosensor; as well as encoding region for Metal Binding Peptide (BBa_K346004) to build a bioaccumulator; and encoding region to mercury reductase (BBa_K1355000).
Source
This construction is based on sequences present in the pO26-CRL plasmid found in Escherichia coli O26. We added a reverse tryptophan operon terminator to ensure transcription termination of the merR messenger RNA. However we didn’t add transcription terminator after the merP gene, aiming connect this biobrick with others, enabling various functions related to mercury (eg. Bio-sensor, bio-remediator, bio-accumulator)
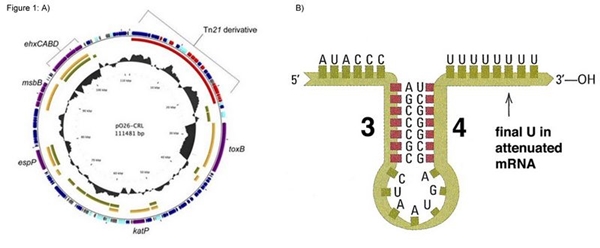
Figure 1: A) p26-CRL plasmid map; B) Illustration of the tryptophan operon terminator.
References
Barkay, T., Miller, S. M., & Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS microbiology reviews, 27(2‐3), 355-384.
BIONDO, R. Engenharia Genética de Cupriavidus metallidurans CH34 para a Biorremediação de efluentes contendo Metais Pesados. 2008. São Paulo, Brasil.
Nascimento, A. M., & Chartone-Souza, E. (2003). Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genetics and Molecular Research, 2(1), 92-101.
DASH, H. R.; DAS, S. Bioremediation of mercury and the importance of bacterial mer genes. 2012. International Biodeterioration & Biodegradation 75: 207 – 213.
HAMLETT, N. V.; et al. Roles of the Tn21 merT, merP and merC Gene Products in Mercury Resistance and Mercury Binding. 1992. Journal of Bacteriology 174: 6377 – 6385.
PINTO, M. N. Bases Moleculares da resistência ao Mercúrio em bactérias gram-negativas da Amazônia brasileira. 2004. Pará, Brasil.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 988
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 586
Illegal NgoMIV site found at 1160 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 579
| None |