Part:BBa_K1326001
ChlM
Mg protoporphyrin IX S-adenosyl methionine O-methyl transferase - Magnesium-protoporphyrin O-methyltransferase (ChlM) [PMID: 12828371; 12489983; 4436384]; ChloroP 1.1 predicts cp location
Operon Usage
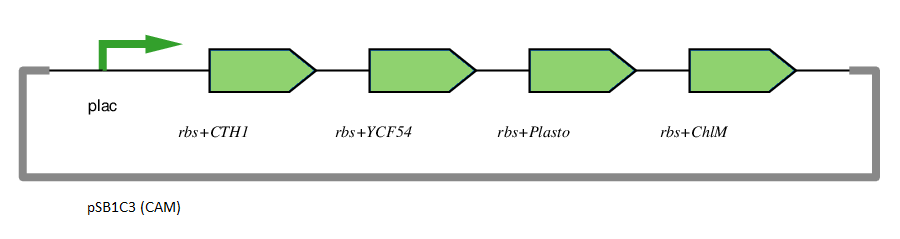
This gene has been used in an operon with other genes responsible for catalysing the biosynthesis pathway from Mg-protoporphyrin IX to Protochlorophyllide. CTH1, Plastocyanin, and YCF54 are involved in the oxidative cyclase pathway. ChlM methylates Mg-protoporphyrin IX, facilitating the highly-regulated catalysis of Mg-chelatase. CTH1 catalyses the conversion of Mg-protoporphyrin IX monomethyl into divinyl protochlorophyllide, interacting with YCF54 and Plastocyanin.
The insertion of magnesium is the key component of the chlorophyll biosynthesis pathway.
The plasmid is under the control of the lac promoter.
Structure
Figure 1: Crystal structure of ChlM from cyanobacteria Synechocystis sp. [http://www.ncbi.nlm.nih.gov/pubmed/25077963]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 230
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 787
- 1000COMPATIBLE WITH RFC[1000]
| None |

