Part:BBa_B0015:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_B0015
- PCR Problems: Using primers VR and VF2 to PCR B0010 results in excess bands. VR can anneal to B0010, resulting in shorter bands than expected. A full description of the problem is available here.
- We used this piece to terminate many of our Berkeley iGEM 2005 Parts. It appeared to work quite well. [Smelissali 6/7/06]
- There have been reports from the Davidson iGEM team that the plasmid from the iGEM 2006 Repository for this part did NOT contain this biobrick. If you are experiencing this or a similar concern, please contact either myself or [http://2006.igem.org/User:Meaganl Meagan Lizarazo] [--Smelissali 14:26, 2 August 2006 (EDT)]
Issues in using this as vector
- The standard assembly described here gives very poor results when whatever part BBa_Xxxxx is to be put in front of BBa_B0015 (< 5 cfu/plate). The procedure described below gave much better results (> 200 cfu/plate):
- Cut BBa_B0015 with XbaI and PstI.
- Cut BBa_Xxxxx with SpeI and PstI.
- Ligate the two parts.
- Bmoeyaert 08:50, 4 September 2008 (UTC)
User Reviews
UNIQ7b5e359efe1b5017-partinfo-00000000-QINU
|
••••
iGEM13_Fudan |
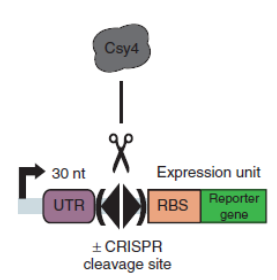
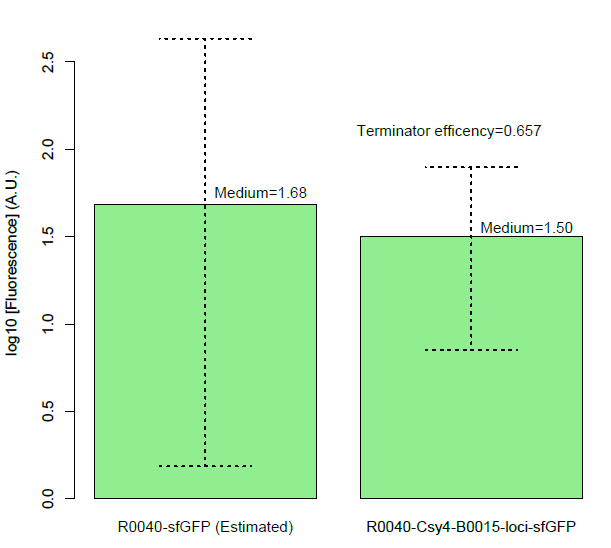
BackgroundCsy4 is a member of CRISPR pathway discovered in Pseudomonas aeruginosa. It is a single endoRNase that recognizes and cleaves a 28-nucleotide repetitive sequence and produces stable transcripts with a 5’hydroxylgroup, which can eliminate unwanted interactions between 5’UTRs and translational elements such as RBSs units to standardize the expression of the elements. ( Lei Qi et al, 2012).The cleavage site can be curtailed to 16nt proved by in vitro data (Rachel E. Huarwitz et al, 2010) Figure. 1. The CRISPR RNA-processing system facilitate engineering of standard genetic elements in various contexts( Lei Qi et al, 2012). DesignCsy4 system is introduced into the terminator efficiency test assay and used optimized sfGFP to test the PoPs of Ptet (BBa_R0040), to approximately estimate the input of BBa_B0015. Then we tested the output of BBa_B0015, with Csy4 insulator to avoid the RNA interference. Figure A. Construction for termination efficiency estimation. We used optimized sfGFP to test the PoPs of Ptet (BBa_R0040), to approximately estimate the input of BBa_B0015. Then we test the output of BBa_B0015, with Csy4 insulator to avoid the RNA interference to sfGFP translation. Experimental Data (Quantitative Measurement)We get the device measured by flow cytometer by testing the expression of sfGFP. Figure B, Histogram of the expression of output of terminator, measured by sfGFP fluorescence by flow cytometer. We assembled the promoter BBa_R0040 and the Csy4 protein with the terminator BBa_B0015. And we used the sfGFP protected by a insulator to measure the leakage rate of the terminator. Then, we found a decrease of terminator efficiency to only 0.657 (Figure C), due to the downstream interference, largely inconsistent with the previous study that showed that BBa_B0015 is a strong terminator, with 0.984[CC], 0.97[JK],forward efficiency, of which the rrnB_T1 (B0010) part also has 97.27% forward efficiency (G. Cambray, et al., 2013). Figure C, Measurement of BBa_B0015 efficiency: column of the median values of fluorescent expressions with/without the terminator and the interaction between the csy4 and the 16-nt cleavage site. (Left: Input/Right: Output) The error bar represents two quartiles.(Up:75%, Down:25%) The terminator efficiency is (1-output value/input value).
Reference: Haurwitz R E, Jinek M, Wiedenheft B, et al. Sequence-and structure-specific RNA processing by a CRISPR endonuclease[J]. Science, 2010, 329(5997): 1355-1358. Qi L, Haurwitz R E, Shao W, et al. RNA processing enables predictable programming of gene expression[J]. Nature biotechnology, 2012, 30(10): 1002-1006. Cambray G, Guimaraes J C, Mutalik V K, et al. Measurement and modeling of intrinsic transcription terminators[J]. Nucleic acids research, 2013, 41(9): 5139-5148. |
Antiquity |width='60%' valign='top'| This review comes from the old result system and indicates that this part worked in some test. |}
|
••••
Aberdeen_Scotland 2009 |
The transformation, Plasmid miniprep and gel (undigested and digested with Xbal and PstI) worked as expected. |
|
•••••
Michigan iGEM 2012 |
As previously observed by other teams, when BBa_B0015 is PCR amplified using VF2 and VR unexpected bands of varying lengths (a bright band at 500bp and several dim ones below 500bp) appear on the electrophoresis gel. |
UNIQ7b5e359efe1b5017-partinfo-00000005-QINU
Background
Csy4 is a member of CRISPR pathway discovered in Pseudomonas aeruginosa. It is a single endoRNase that recognizes and cleaves a 28-nucleotide repetitive sequence and produces stable transcripts with a 5’hydroxylgroup, which can eliminate unwanted interactions between 5’UTRs and translational elements such as RBSs units to standardize the expression of the elements. ( Lei Qi et al, 2012).The cleavage site can be curtailed to 16nt proved by in vitro data (Rachel E. Huarwitz et al, 2010)
Figure. 1. The CRISPR RNA-processing system facilitate engineering of standard genetic elements in various contexts( Lei Qi et al, 2012).
Design
Csy4 system is introduced into the terminator efficiency test assay and used optimized sfGFP to test the PoPs of Ptet (BBa_R0040), to approximately estimate the input of BBa_B0015. Then we tested the output of BBa_B0015, with Csy4 insulator to avoid the RNA interference.
Figure A. Construction for termination efficiency estimation. We used optimized sfGFP to test the PoPs of Ptet (BBa_R0040), to approximately estimate the input of BBa_B0015. Then we test the output of BBa_B0015, with Csy4 insulator to avoid the RNA interference to sfGFP translation.
Experimental Data (Quantitative Measurement)
We get the device measured by flow cytometer by testing the expression of sfGFP.
Figure B, Histogram of the expression of output of terminator, measured by sfGFP fluorescence by flow cytometer. We assembled the promoter BBa_R0040 and the Csy4 protein with the terminator BBa_B0015. And we used the sfGFP protected by a insulator to measure the leakage rate of the terminator.
Then, we found a decrease of terminator efficiency to only 0.657 (Figure C), due to the downstream interference, largely inconsistent with the previous study that showed that BBa_B0015 is a strong terminator, with 0.984[CC], 0.97[JK],forward efficiency, of which the rrnB_T1 (B0010) part also has 97.27% forward efficiency (G. Cambray, et al., 2013).
Figure C, Measurement of BBa_B0015 efficiency: column of the median values of fluorescent expressions with/without the terminator and the interaction between the csy4 and the 16-nt cleavage site. (Left: Input/Right: Output) The error bar represents two quartiles.(Up:75%, Down:25%) The terminator efficiency is (1-output value/input value).
Reference:
Haurwitz R E, Jinek M, Wiedenheft B, et al. Sequence-and structure-specific RNA processing by a CRISPR endonuclease[J]. Science, 2010, 329(5997): 1355-1358. Qi L, Haurwitz R E, Shao W, et al. RNA processing enables predictable programming of gene expression[J]. Nature biotechnology, 2012, 30(10): 1002-1006. Cambray G, Guimaraes J C, Mutalik V K, et al. Measurement and modeling of intrinsic transcription terminators[J]. Nucleic acids research, 2013, 41(9): 5139-5148.
</I> |width='60%' valign='top'| Enter the review inofrmation here. |}; Antiquity |width='60%' valign='top'| This review comes from the old result system and indicates that this part worked in some test. |}
|
••••
Aberdeen_Scotland 2009 |
The transformation, Plasmid miniprep and gel (undigested and digested with Xbal and PstI) worked as expected. |
|
•••••
Michigan iGEM 2012 |
As previously observed by other teams, when BBa_B0015 is PCR amplified using VF2 and VR unexpected bands of varying lengths (a bright band at 500bp and several dim ones below 500bp) appear on the electrophoresis gel. |
UNIQ7b5e359efe1b5017-partinfo-00000009-QINU

 1 Registry Star
1 Registry Star