Part:BBa_K4192021
obcB, one of the genes required for oxalic acid synthesis
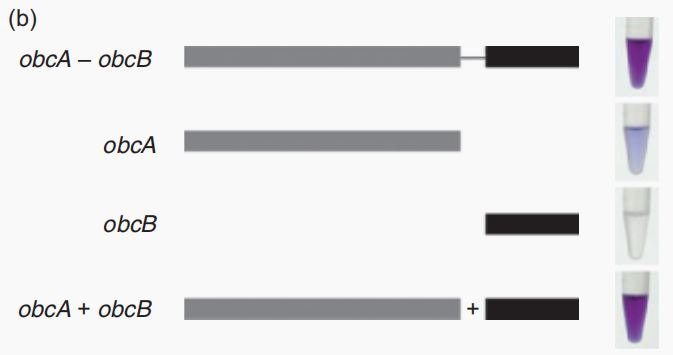
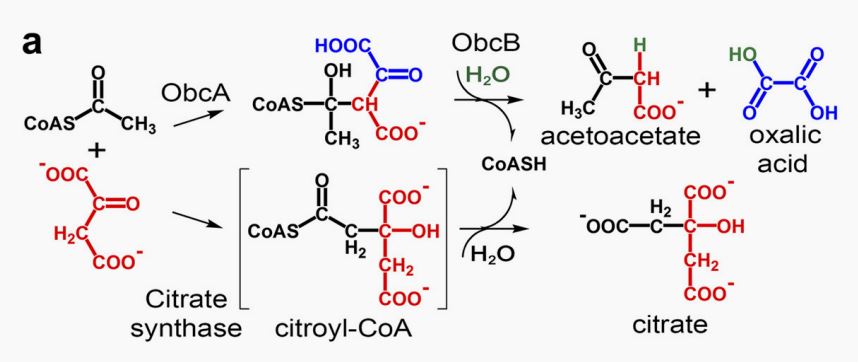
It is one of the genes required for oxalic acid synthesis in Burkholderia glumae, which can act together with ObcB to catalyze oxaloacetic acid (OAA) and citric acid to form oxalic acid (OA). When there is only ObcB, it cannot form oxalic acid.
Biology
CAU_China 2022 used this part in their oxalate secretion circuit and the expression product of obcB is one of the main enzyme in in the oxalate secretion circuit[1]. It can act together with ObcA to catalyze oxaloacetic acid (OAA) and citric acid to form oxalic acid (OA). When there is only ObcB, it cannot form oxalic acid[2].
Fig.1 Effects of obcA and obcB, the shade of purple reflects the concentration of oxalic acid[1].
When producing oxalic acid, obcB turns C6-CoA adduct into CoA, acetoacetate,and oxalic acid
Fig.2 Mechanism of obcA and obcB[2].
Considering the importance of obcA and obcB occur togrther in the biosynthesis of oxalic acid, the CDS of them are in close proximity to each other, even encoded on a single polycistronic message. The implication of this for our project is that we can make these two genes share the same promoter[2].
Fig.3 Distribution of obcA and obcB[1].
We constructed a separate pET-obcB plasmid, and determined that the ObcB protein can be successfully expressed in E.coli BL21 (DE3) through SDS-PAGE, the results are as follows:
Fig.4 Obc protein in SDS-PAGE.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 669
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 669
Illegal NotI site found at 582
Illegal NotI site found at 687 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 650
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 669
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 669
Illegal NgoMIV site found at 460
Illegal NgoMIV site found at 501
Illegal NgoMIV site found at 625 - 1000COMPATIBLE WITH RFC[1000]
References
[1]Nakata, Paul A, and Cixin He. “Oxalic acid biosynthesis is encoded by an operon in Burkholderia glumae.” FEMS microbiology letters vol. 304,2 (2010): 177-82. doi:10.1111/j.1574-6968.2010.01895.x
[2]Oh, Juntaek et al. “Structural basis for bacterial quorum sensing-mediated oxalogenesis.” The Journal of biological chemistry vol. 289,16 (2014): 11465-11475. doi:10.1074/jbc.M113.543462
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 489
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 367
Illegal NgoMIV site found at 408 - 1000COMPATIBLE WITH RFC[1000]
| None |