Part:BBa_K2086002
PyeaR - SerA This part is a composite part of the promoter BBa_K216005 with the serine coding sequence BBa_K2086002 as well as a standard RBS BBa_B0030 and terminator BBa_B0015. It is designed to act as a nitrate sensitive kill-switch by utilizing a nitrate sensitive promoter coordinated with an essential gene to complement an auxotrophic strain of E. coli only under high nitrate conditions.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 422
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Biology and Our Application
PyeaR
The yeaR-yoaG operon is found in E. coli and has been shown to be sensitive to nitrate concentrations as well as nitrite concentrations to a lesser degree [1]. The promoter of this operon is regulated by the Nar and NsrR repressors found. These repressors are deactivated in the presence of nitrate,nitrite, and nitric oxide to a lesser extent. This promoter is also unique in the fact that it is not repressed under aerobic conditions. The original BioBrick BBa_K216005 was made by the 2009 Edinburgh iGEM team.
[1] HSIA-YIN LIN, PEGGY J. BLEDSOE, AND VALLEY STEWART: Activation of yeaR-yoaG Operon Transcription by the Nitrate-Responsive Regulator NarL Is Independent of Oxygen- Responsive Regulator Fnr in Escherichia coli K-12 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2168752/
Serine
Serine is an amino acid produced in E. coli K12 through the metabolic pathway shown in Figure 2. The SerA gene codes for D-3-Phosphoglycerate Dehydrogenase, the enzyme responsible for catalyzing the committed step of serine biosynthesis. Without SerA, E. coli are unable to grow without sufficient supplementation of other amino acids [2].
Taking advantage of the bacteria's dependence on serine, we planned to create a safety kill switch by controlling the production of the amino acid. By obtaining an auxotrophic strain missing the SerA strain (JW2880 [3]) we were able to create a complement SerA plasmid to rescue the strain when grown in media without supplementary amino acids. Our kill-switch (BBa_K2086002) was incorporated into our final project design.
[2] PAULA D. RAVNIKAR AND RONALD L. SOMERVILLE: Genetic Characterization of a Highly Efficient Alternate Pathway of Serine Biosynthesis in Escherichia coli. http://jb.asm.org/content/169/6/2611.full.pdf
[3] Coli Generic Stock Center: JW2880. http://cgsc.biology.yale.edu/Strain.php?ID=108515
SerA Supplementation
Before interpreting the results from our composite part including SerA, it's important to first consider whether our selected auxotroph (JW2880) was able to be rescued with a supplemental SerA gene. By complementing our auxotroph with a plasmid containing SerA and its native promoter, the growth curve in Figure 3 was obtained. After this experiment, we can safely conclude that our auxotrophic strain is able to complemented with a serA plasmid.
PyeaR-serA
The ability of our full composite part to rescue our auxotroph was confirmed by testing our composite BioBrick kill switch (BBa_K2086002), which expresses the SerA gene when nitrate concentrations reach a certain threshold. The qualitative results from one of these experiments are pictured in Figure 4. In the solution with high nitrate concentration, the cells were able to survive, while the culture in minimal medium was not able to grow.

2022 NEU_CHINA
Part BBa_K4220100 is an improvemnet of this part.
Why is this an improvement?
We have improved a kill switch in E. coli (composite part) into one which can function in the yeast by replacing each basic parts into ones which have similar functions in the yeast.
Changes:
- Promoter: PyeaR promoter (responsive to nitrate, nitrite and nitric oxide) to CUP1 (responsive to copper ion)
- Essential gene: SerA (E. coli) to HSF1 (yeast)
Design
- How to kill the yeast?
- HSF1: essential gene in yeast [1]
- How to determine when to kill?
- Copper ion: abundant in the intestine; scarce in the environment [2]
- Others
- HIS3-NatR-ADH1: resistant gene expression for screening
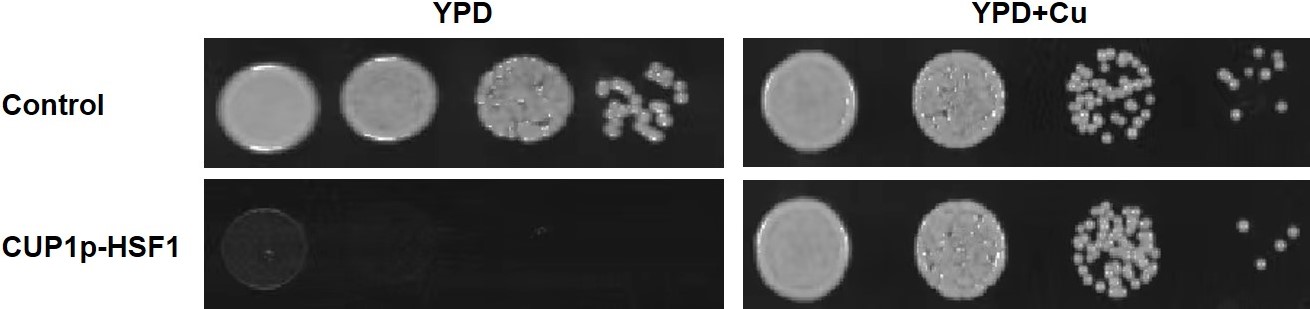
Results
CUP1p-HSF1: kill switch
YPD: Yeast Extract Peptone Dextrose Medium
| None |