Part:BBa_K3739114
K525998-Aly01-his-3SpyCatcher-B0010
T7-Aly01-his-3SpyCatcher
Biology
SpyCatcher
SpyTag (a 13-residue peptide) and SpyCatcher (a 116-residue domain) are engineered split fragments of the second immunoglobulin-like collagen adhesin domain (CnaB2) of the fibronectin-binding protein (FbaB) of Streptococcus pyogenes. iGEM team Peking 2016 has modified the peptide and domain into triple SpyTag and triple SpyCatcher, which can form bonds with each other and create Spy Crosslinking Network, enabling the formation of hyper-branched products and polymer network at low concentrations, and gels with a high degree of cross-linkage at high concentrations.
Aly01
Aly01 is alginate lyase from Vibrio natriegens SK42.001, which is secreted out and able to digest alginate to unsaturated alginate oligosaccharide. Its signal peptide (named Aly01 in our parts), which is fused with heterogenous protein, is performed well in E. coli. It is implied that the heterogenous protein fused with Aly01 signal peptide may be also secreted efficiently.
Usage
In order to get secreted 3SpyCatcher and further purify it, we added the signal peptide Aly01, followed by a his-tag (6*His) at the N-terminal of 3SpyCatcher. We used both BBa_K081005 and BBa_K525998 to construct the expression system and obtained the composite BBa_K3739034 and BBa_K3739114, which are assembled on the expression vector pET-28a(+) by standard assembly. The constructed plasmids were transformed into Vibrio natriegens, then the positive transformants were selected by kanamycin and confirmed by colony PCR and sequencing.
Characterization
1. Agarose Gel Electrophoresis
After transferring the plasmid into Vibrio natriegens, colony PCR was used to certify the plasmid was correct. The expected result was obtained(2101bp).
Fig.1 The result of colony PCR. Plasmid pET-28a (+).
Characterization of signal peptides
Since some proteins in our project are intended to function in extracellular environment, how to get out of the cells is vital for realization of their functions. Signal peptide is a short amino acid sequence on secreted protein, which is a natural helper of protein to get out of cytoplasm. However, signal peptides can lead protein go to not only the extracellular environment but also the periplasm and membrane. There are several signal peptides selected for V.natriegens, but these are all directed into periplasm. 1 Although there is a signal peptide of alginate lyase from V.natriegens able to direct heterogenous protein to extracellular environment in E.coli chasis, It is not clear that there is a signal peptide conducting protein to extracellular environment in V.natriegens. 2
Selected procedure
Here, we use a tip to select the signal peptide. Download the translated CDS information of V.natriegens from NCBI Assembly. Through analysis of SignalP 5.0, we found out several candidate signal peptides which have significant likelihood to work with corresponding secretion system. According to the function of proteins containing proposed signal peptide, we can exclude the signal peptides targeting to the periplasm and cell membrane. Finally, we find out the best candidate signal peptide, LMT signal peptide, from our chassis
Qualification results
To clarify whether the signal peptides we selected are able to direct the fused proteins to extracellular environment, we constructed Aly01 and LMT single peptides with BBa_K2560063 which is from 2018 Marhurg iGEM program and transformed the pET28a(+) plasmid with composite parts BBa_K3739038 and BBa_K3739041 into V.natriegens. Then we use the anti-GFP antibody to detect secreted GFP in the filtered supernatant of V.natriegens liquid culture by western blotting after V.natriegens is cultivated for .16 hours. The concentration of his-GFP in the supernatant of BBa_K3739041 is significantly higher than that in J23100-B0030-GFP. (Fig. 1A, 1B) Furthermore, less expression of BBa_K3739041 than that of GFP conforms that LMT-his-GFP is secreted rather than from lysed cells. (Fig. 1C) However, the concentration of Aly01-his-GFP is less than that of GFP. (Fig. 1A, 1B) It is probably because that there are less expression of Aly01-his-GFP in V.natriegens. (Fig. 1C) Thus, we use another method to prove our hypothesis.
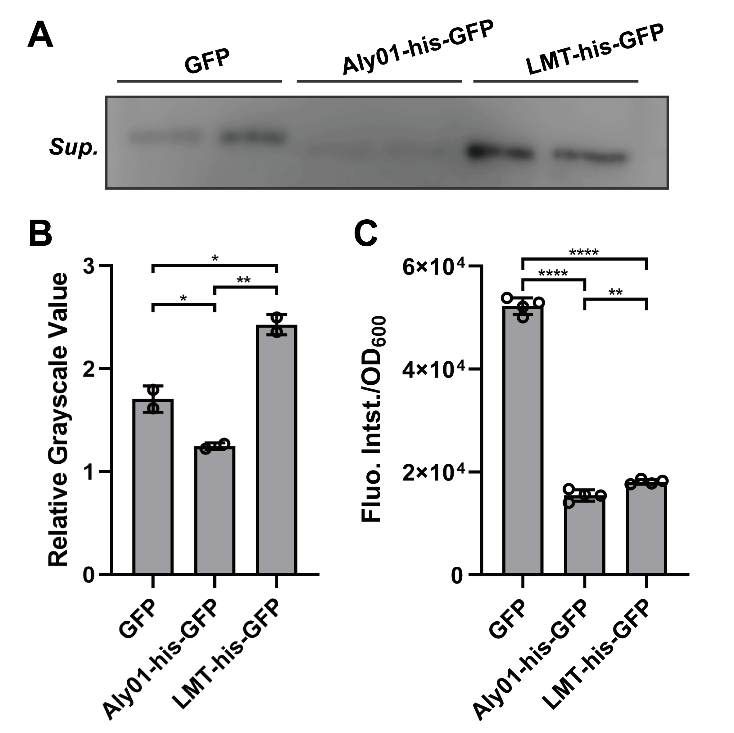
Fig. 1. (A) Western Blotting detection of the V.natriegens supernatants of J23100-B0030-GFP, BBa_K3739038 and BBa_K3739041 after 16h cultivation in LBv2 liquid culture, 37℃. Sup. means supernatant. (B) Grayscale analysis of (A). (C) Fluorescence intensity/OD600 value of V.natriegens strain of J23100-B0030-GFP, BBa_K3739038 and BBa_K3739041 in LBv2 liquid culture after 16h cultivation in 37℃.
Quantitation results
In order to exclude the difference of expression among J23100-B0030-GFP, BBa_K3739038 and BBa_K3739041 and quantify secretion effect of signal peptide, we treated V.natriegens liquid cultures after 12.5 h cultivation in 37℃ with 12.5 μg/mL chloramphenicol. Then we sampled the cultures every 6 minutes and terminated secretion with 20 mM NaN3. As time goes, the his-GFP concentration of culture supernatant were increased in both BBa_K3739038 and BBa_K3739041. This is indicated that both LMT and Aly01 signal peptide are able to translocate fused protein to extracellular environment. (Fig. 2A, 2B) We made a grayscale analysis for the modelling of secretion, and the total GFP is detected for confirmation that the translation is completely repressed. (See our Model https://2021.igem.org/Team:XMU-China/Model pages)
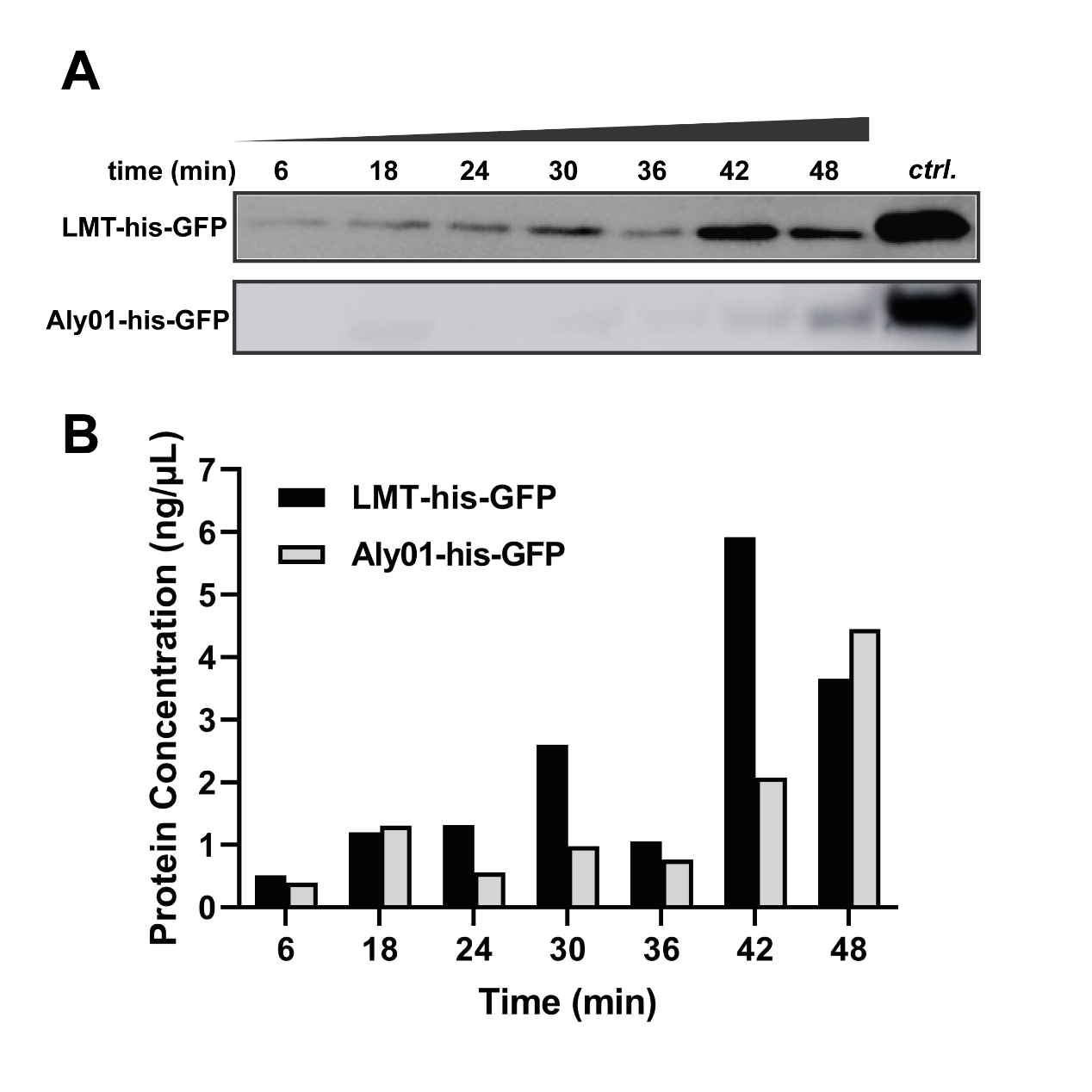
Fig. 2. (A) Western blotting detection of sampled supernatants treated with chloramphenicol in different times. (B) GFP concentration of treated supernatant of BBa_K3739038 and BBa_K3739041 in (A)
Secretion in E.coli
Besides conducting protein secretion in V.natriegens, Aly01 signal peptide is reported that it is able to conduct protein secretion in E.coli as well. 2We supposed that LMT signal peptide may also have conducting function in E.coli. Thus, we used filtered culture of BL21 strain transformed with pET28a(+)-BBa_K3739043 and pET28a(+)-BBa_K3739041 for SDS-PAGE. Then the SDS-PAGE gel is stained by silver staining. Contrasting to BBa_K3739043, his-GFP is detected in the supernatant of pET28a(+)-BBa_K3739041.(Fig. 3) It is indicated that LMT signal peptide can also work for conducting ====secretion in E.coli====
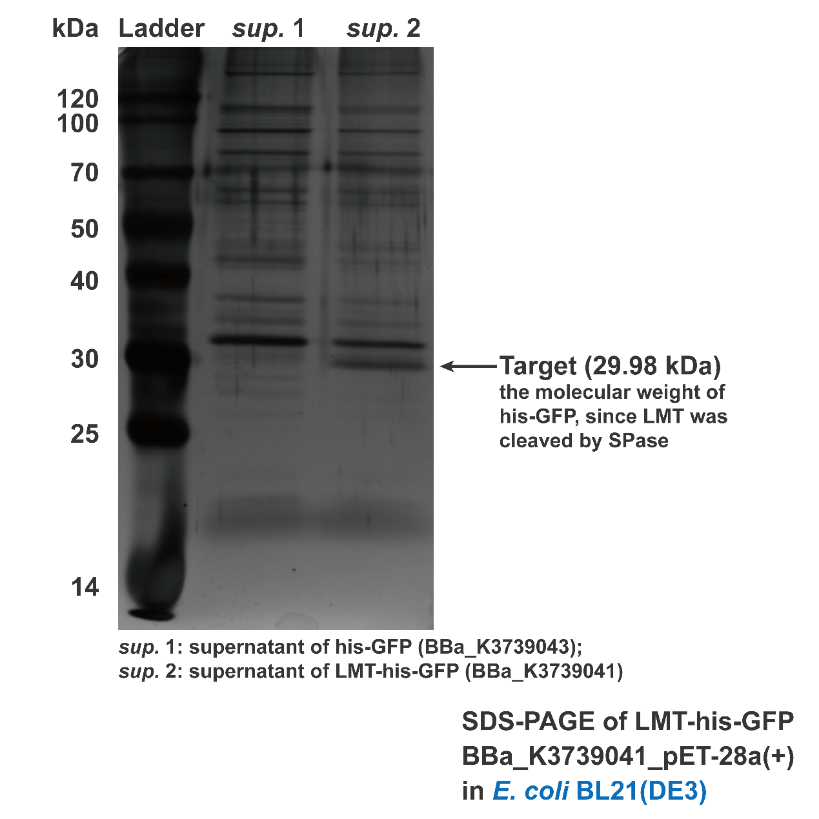
Fig. 3. Silver staining result of supernatant of BL21(DE3) transformed with BBa_K3739043 and BBa_K3739041, respectively.
Then we used an alternative way to verify the secretion function of signal peptides in E.coIi. The proteins labeled of tetracysteine motif tag (FlAsH tag, FT in short) could be detected with the biarsenical compound FlAsH-EDT2. 3
To quantify and qualify the secretion effect, FT was added into the C-terminal of BBa_K3739000, BBa_K3739006 and BBa_K3739009. Composite parts BBa_K3739084, BBa_K3739085, BBa_K3739087 were constructed (Fig. 4) on plasmid backbone pSB1C3 or pET-28a(+) and then the recombinant plasmid was transformed into E. coli BL21 (DE3). Positive colonies selected by colony PCR and sequencing were cultivated for secretion analysis.
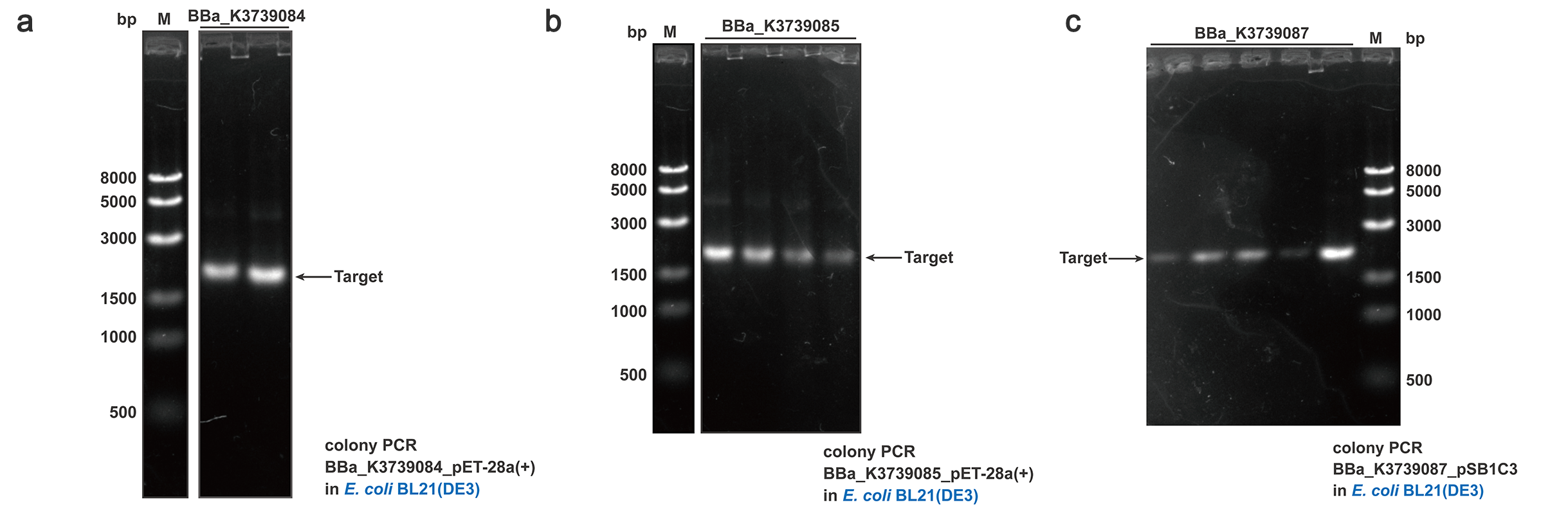
Fig. 4. Colony PCR results of BBa_K3739084, BBa_K3739085, BBa_K3739087
After cultivation for 5 hours at 37℃ and induction for 2 hours with 0.1 mM IPTG, the cell supernatant was separated by centrifugation. The supernatant along with 2 μM FlAsH-EDT2 and 1 mM DTT were added to wells in a 96-well plate. Following incubation in the dark for 1 h at 37 °C, fluorescence was measured by 503 nm excitation and 528 nm emission, and each value was normalized by OD600. The result shows that the fluorescence intensity/OD600 of the group Aly01 and LMT is significantly higher than native control group, which proves that these 2 signal peptides can function well. (Fig. 5)
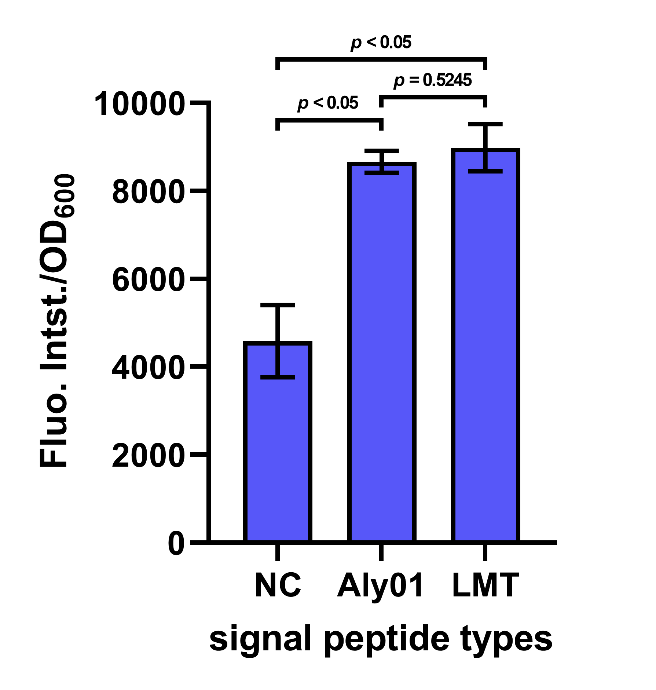
Fig. 5. Fluorescence intensity/OD600 of cell supernatant after incubation in the dark for 1h.
Moreover, to promote our understanding on secretion process mediated by signal peptides, we built a model about relationship between the amount of secreted recombinant protein and time. (See our Model https://2021.igem.org/Team:XMU-China/Model pages) In order to get the value of P_T (the total amount of recombinant protein per unit volume of reactor), P_M (the amount of the recombinant protein in the medium compartment in unit volume) and γ(extracellular secretion rate constant), a sets of characterization experiments were done.
BBa_K3739087 were constructed on plasmid backbone pET-28a(+) and then the recombinant plasmid was transformed into E. coli BL21 (DE3). After cultivation for 5 hours and induction for 3 hours with 0.1 mM IPTG, 50mg/L chloroamphenicol was added to bog down the synthesis of protein in the engineered bacteria in order to make the experimental operations more convenient. Then, the cell supernatant was collected in 0, 6, 12, 18, 24, 30, 40 min to test the amount of secreted recombinant protein by fluorescence detection of FlAsH-EDT2 and FT. (Fig. 6)
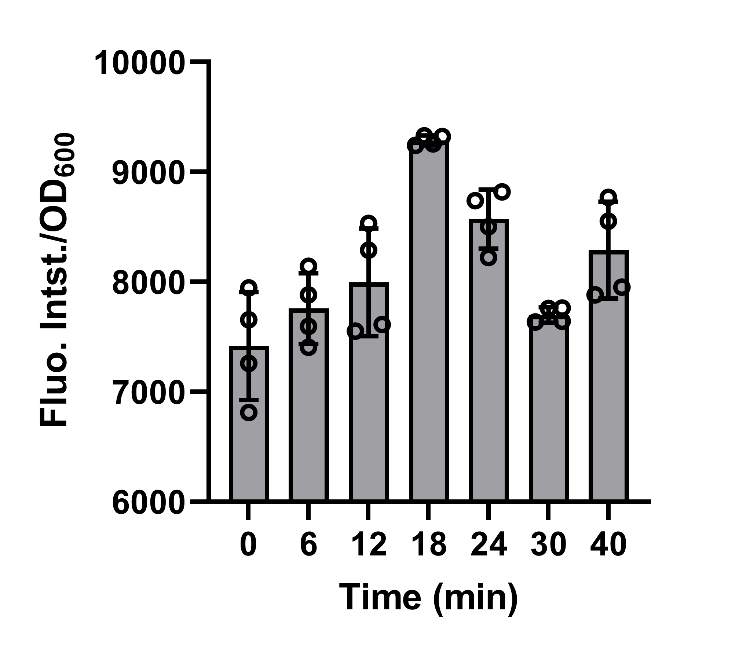
The data shows that after the protein synthesis was inhibited, the change in the amount of secreted protein over time was well matched with the model.
Reference
1. Yang, X.; Wei, J.; Wang, Y.; Yang, C.; Zhao, S.; Li, C.; Dong, Y.; Bai, K.; Li, Y.; Teng, H.; Wang, D.; Lyu, N.; Li, J.; Chang, X.; Ning, X.; Ouyang, Q.; Zhang, Y.; Qian, L., A Genetically Encoded Protein Polymer for Uranyl Binding and Extraction Based on the SpyTag-SpyCatcher Chemistry. ACS Synth Biol 2018, 7 (10), 2331-2339.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1464
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 177
| None |

