Part:BBa_K2148016
C.reinhardtii chloroplast rbcL 3UTR
This part contains the 3'UTR for the rbcL gene found in Chlamydomonas reinhardtii chloroplast genome. This is coded as a level-0 3UTR/TERM Phytobrick, which together with other level-0 Phytobrick can form a desired transcriptional unit for transformation.
Usage and Biology
This is a level-0 3UTR/TERM Phytobrick. It can be used along with other level-0 Phytobrick promoters and CDS to create a transcriptional unit.
Caution: In chloroplast rbcL 3'UTR don't serve as transcription terminators as in bacteria, but as elements for transcript maturation/stabilisation. atpB 3'UTR can be used as an alternative. (on advice from Prof. Saul Purton - UCL)
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 490
Illegal XbaI site found at 478
Illegal XbaI site found at 520
Illegal SpeI site found at 514
Illegal PstI site found at 28
Illegal PstI site found at 496 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 490
Illegal NheI site found at 42
Illegal SpeI site found at 514
Illegal PstI site found at 28
Illegal PstI site found at 496
Illegal NotI site found at 526 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 490
Illegal BamHI site found at 508 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 490
Illegal XbaI site found at 478
Illegal XbaI site found at 520
Illegal SpeI site found at 514
Illegal PstI site found at 28
Illegal PstI site found at 496 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 490
Illegal XbaI site found at 478
Illegal XbaI site found at 520
Illegal SpeI site found at 514
Illegal PstI site found at 28
Illegal PstI site found at 496 - 1000COMPATIBLE WITH RFC[1000]
Characterisation
iGEM Marburg 2021 - Improvement of an Existing Part
Motivation
Cell-free technology offers the possibility for high-throughput characterization of genetic parts in a shorter time frame. This timecut is notable when working with plants in general, but is even more of a hurdle when working on the chloroplast. Normally, it takes several months after a genetic device is introduced until it can be qualitatively/quantitatively characterized. The integration of a transgene in the chloroplast of an organism of choice includes very costly lab equipment and consumables. On top of this, the methods do not offer support to test multiple genetic devices in a time-efficient manner and for many plant species, no chloroplast transformation protocols are available to date.
Currently, the repertoire of available genetic parts in the field of chloroplast plant synthetic biology is hugely limited and the characterisation of these tools is very cost-intensive and time consuming. Although a handful of inducible parts and regulatory sequences exist [3] [4] [5] [6], the small collection poses a huge hurdle for more complex engineering projects. In literature, a lot of ambitious and aspirational projects have been proposed like the implementation of nitrogen fixation pathways into plants or increasing nutritional value in plants [7] [8]. But when considering the limited availability of parts, projects like these seem impossible at the current time.
Our Part of Choice
The iGEM registry is one of the cornerstones of synthetic biology as it encompasses a collection of over 20.000 parts. As it is normal for iGEM projects to not be completely finished until the end of the iGEM season, we aimed to connect to the efforts of [http://2016.igem.org/Team:Cambridge-JIC iGEM Cambridge 2016]. They introduced a chloroplast part collection for the model algae Chlamydomonas reinhardtii. The toolbox included regulatory sequences, reporter genes, a CRISPR/Cas9 system and a set of homology flanks for genomic integration. As we think it is of high priority to connect to older iGEM projects and reiterate designs during future projects, we decided to have a deeper look into their parts. During a closer inspection we noticed that the 3’UTR of the rbcL gene (large subunit of the RuBisCo gene) BBa_K2148016 BioBrick of their collection was falsely linked to one of their other parts BBa_K2148007. Shortly after, we informed Vinoo Selvarajah head of the iGEM registry about this and consequently the part’s annotation has been corrected.
Analysis of the existing part
When having a closer look at the part sequence, we noticed remnants of earlier cloning steps. Apart from the 3’UTR region of the rbcL, a large portion of the coding sequence and fragments of the original vector have been included in the part[8], which they took from the cloning vector pASapI [9].
According to our definition of synthetic biology: The foundation of synthetic biology lies in applying engineering principles to biology and one of the key aspects for that is standardization, which is especially important for the creation of standardized biological parts.
As we are dedicated to standardization, we wanted to check if the part would still be functional if we only include the rbcL 3’UTR region and if we detect any significant change in expression when we use different lengths of the 3’UTR.
With this in mind we took our 3’UTR measurement construct with which we can easily characterize 3’UTR sequences in a high-throughput manner. We therefore exchanged the inserted 3’UTR dropout cassette - a MoClo compatible GFP placeholder introduced by iGEM Marburg 2019 - with the 3 versions of the 3’UTR of Chlamydomonas reinhardtii and tested them in Tobacco chloroplast cell-free extracts in order to test their viability.
But before conducting the actual experiment we wanted to compare the secondary structure of the different 3’UTRs to see if any significant stem structures could be detected (Figure 1,2,3)
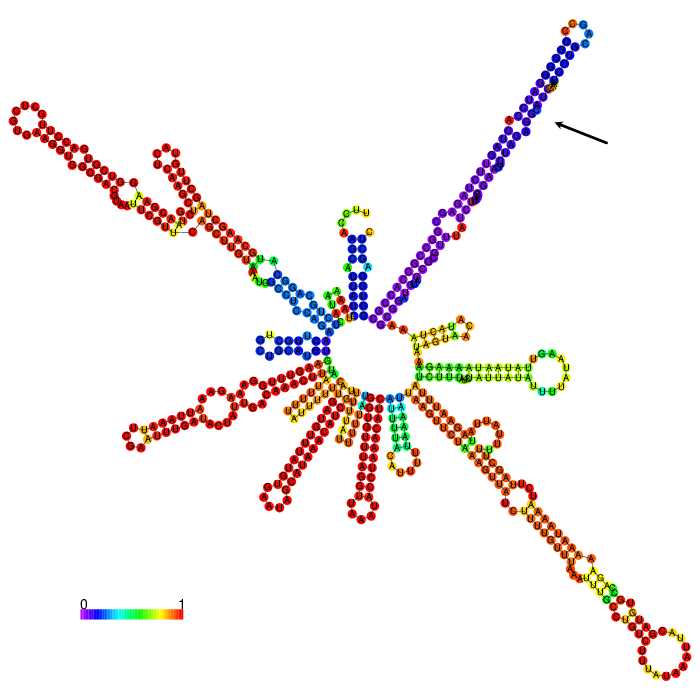
The part forms a complex structure due to its length. At the 3’ end, a long stem loop structure can be seen, which the algorithm could not confidently predict.
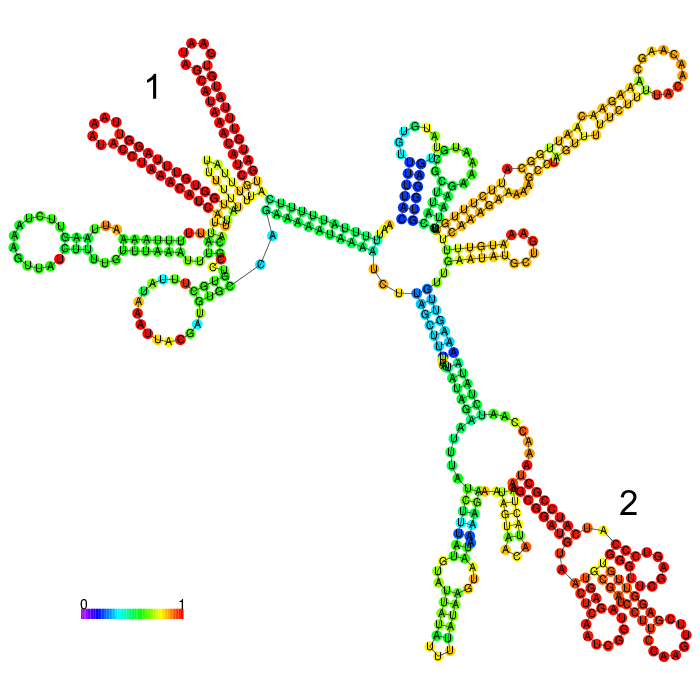
The part forms a complex structure with two strong distinct stem loop structures near the beginning of the 5’ end of the 3’UTR (visible at the top left). Another strong distinct structure can be seen approximately in the middle of the sequence, which is a tRNA we included in the part for potential higher expression (bottom right)
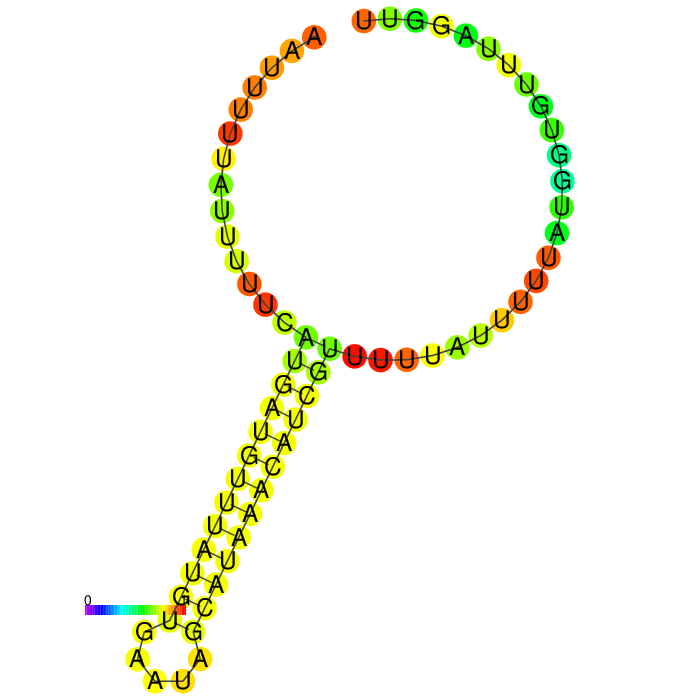
A strong stem loop structure can be detected. This is the short version we designed, because we noticed a huge drop in our Transcriptomic data set directly upstream of this stem loop
Comparison of the 3'UTR versions
3’ untranslated regions in chloroplasts do not necessarily function as efficient transcription terminators. They rather form distinct secondary structures that stabilizes the RNA and protects it from 3’ endonucleases. During our research on the 3’UTR of rbcL from Chlamydomonas reinhardtii we found out that in fact 60% of transcription does not terminate at the 3’UTR region, but instead continues downstream [10]. On top of this we also found out that there are two distinct stem loops found directly downstream the stopcodon of the rbcL gene that are thought to protect the mature RNA from degradation. These stem loops were clearly visible in the long version of the 3’UTR rbcL. In the short version of the rbcL 3’UTR only the first loop structure is visible. This is due to us designing this part in accordance with our Transcriptomic data set that showed a huge drop in reads directly upstream of the first stem loop structure.
After comparing the different secondary structure predictions we noticed that the model had problems accurately predicting the fold of the BioBrick of Cambridge, while our versions were easier to predict for the algorithm. It was also notable that the longer version showed good stem loop structures, because of the tRNA that was included in the part. The smaller version included a potentially strong stem loop structure that is enveloped by two poly-U regions.
With this in mind we went ahead and conducted the actual measurement.
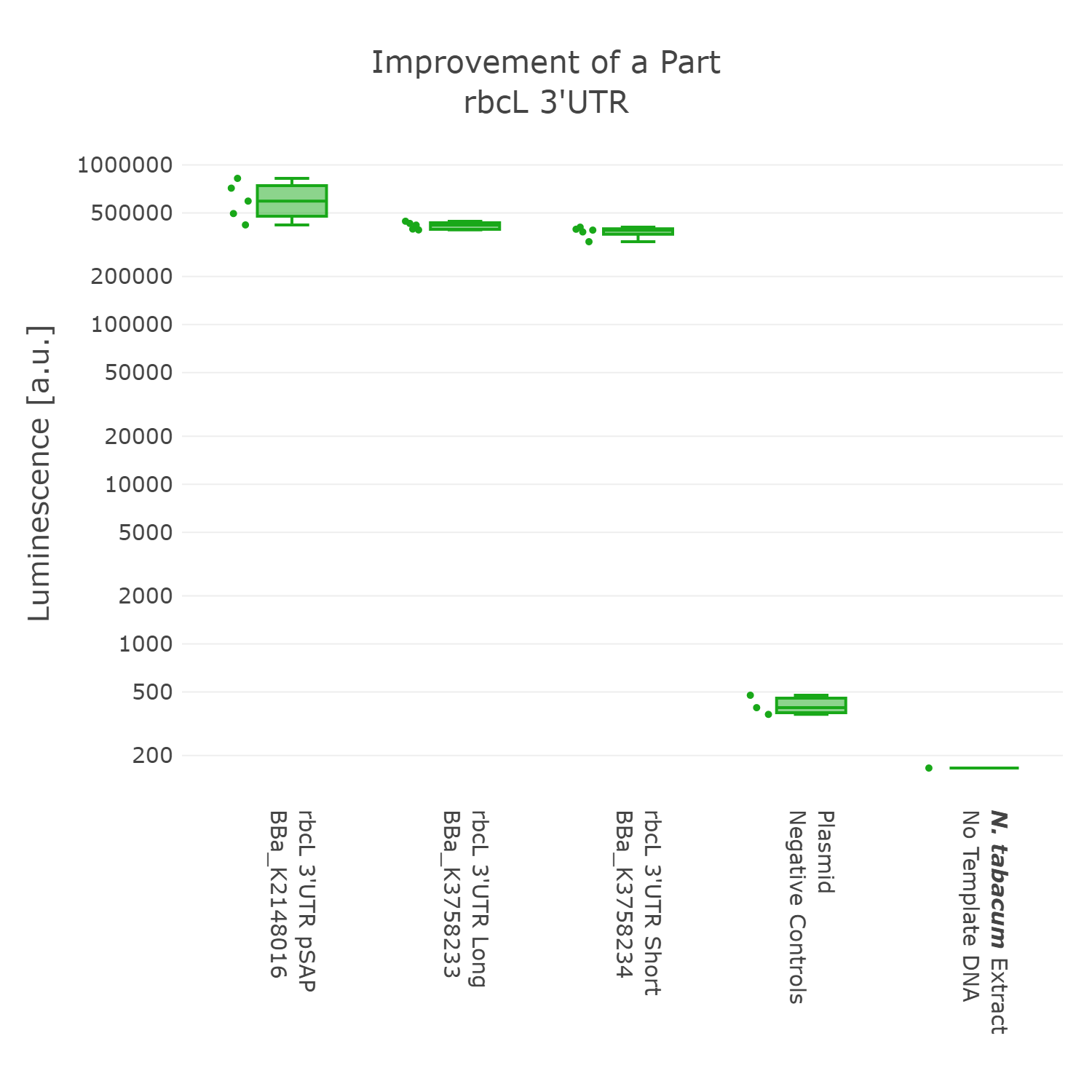
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed with the BioBricks BBa_K2148016, BBa_K3758233 and BBa_K3758234 that were introduced into the 3’UTR measurement lvl2 vector
Conclusion
On the basis of the data we could not detect any significant difference between the three 3’UTR sequences. Although, the original BioBrick performed slightly better. In accordance with our dedication to standardization, we suggest not to include remnants from older cloning projects in parts as they would accumulate over time and could negatively influence genetic designs in the future. Also, in order to better track the history of parts it might be better to only include sequences that are directly graspable from the parts name. We therefore present BBa_K3758234 as our improved version of the rbcL 3’UTR of Chlamydomonas reinhardtii.
References
[1] Wannathong T, Waterhouse JC, Young REB, Economou CK, Purton S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Applied Microbiology and Biotechnology. 2016;100:5467-5477. doi:10.1007/s00253-016-7354-6.
[2] Michel Goldschmidt-Clermont, Miche`le Rahire and Jean-David Rochaix. Redundant cis-acting determinants of 3¢ processingand RNA stability in the chloroplast rbcL mRNA ofChlamydomonas. The Plant Journal (2008) 53, 566–577. doi: 10.1111/j.1365-313X.2007.03365.x
[3] Mühlbauer, S. K., & Koop, H.-U. (2005). External control of transgene expression in tobacco plastids using the bacterial lac repressor. In The Plant Journal (Vol. 43, Issue 6, pp. 941–946). Wiley. https://doi.org/10.1111/j.1365-313x.2005.02495.x
[4] Verhounig, A., Karcher, D., & Bock, R. (2010). Inducible gene expression from the plastid genome by a synthetic riboswitch. In Proceedings of the National Academy of Sciences (Vol. 107, Issue 14, pp. 6204–6209). Proceedings of the National Academy of Sciences. https://doi.org/10.1073/pnas.0914423107
[5] Surzycki, R., Cournac, L., Peltier, G., & Rochaix, J.-D. (2007). Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas. In Proceedings of the National Academy of Sciences (Vol. 104, Issue 44, pp. 17548–17553). Proceedings of the National Academy of Sciences. https://doi.org/10.1073/pnas.0704205104
[6] Rojas, M., Yu, Q., Williams-Carrier, R., Maliga, P., & Barkan, A. (2019). Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. In Nature Plants (Vol. 5, Issue 5, pp. 505–511). Springer Science and Business Media LLC. https://doi.org/10.1038/s41477-019-0412-1
[7] Li, Q., & Chen, S. (2020). Transfer of Nitrogen Fixation (nif) Genes to Non‐diazotrophic Hosts. In ChemBioChem (Vol. 21, Issue 12, pp. 1717–1722). Wiley. https://doi.org/10.1002/cbic.201900784
[8] Goicoechea, N., & Antolín, M. C. (2017). Increased nutritional value in food crops. In Microbial Biotechnology (Vol. 10, Issue 5, pp. 1004–1007). Wiley. https://doi.org/10.1111/1751-7915.12764
| None |
