Part:BBa_K1614000
T7 promoter for expression of functional RNA
Minimal promoter derived from T7 phage promoter.
Usage and Biology
This T7 promoter derivate is especially useful for the use in in vitro applications. The G which is the startpoint for transcription is included in the promoter sequence.
Characterize the T7 promoter – CCU-Taiwan
In our pHMT-LbCas12a construct, we included the T7 promoter (Part:BBa_K1614000) to induce the LbCas12a expression. To characterize the ability of T7 promoter to induce functional RNA, we examined whether the T7 promoter induced LbCas12a RNA can be translated into proteins by SDS-PAGE and Coomassie blue staining. We first transformed pHMT-LbCas12a into E.coli BL21(DE3), which harbors a Lac Operon regulated T7 polymerase. We then expanded the transformed BL21(DE3) cells by TB medium at 37℃. When O.D.600 approach 0.6 ~ 0.8, we induced T7 polymerase expression by adding 0.2 mM IPTG to TB medium and shift BL21(DE3) to 16℃ for 16 hr.
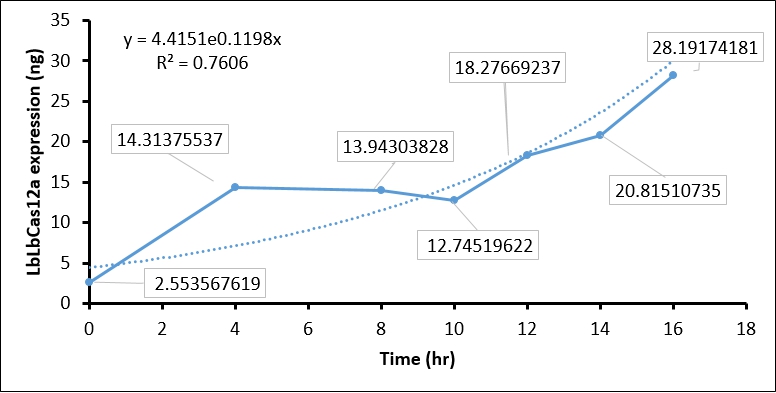
The IPTG treatment increased the T7 polymerase expression in BL21(DE3) as time goes on, which in turn activate T7 promoter. The activated T7 promoter should promote LbCas12 transcription and translation. To examine this, we collected IPTG-induced BL21(DE3) every two hour, and performed SDS-PAGE assay and Coomassie Blue staining to detect LbCas12a protein expression. In brief, 1 ml of BL21(DE3) is harvested every two hour and centrifuged to cell pellet. The cell pellet was then sonicated, and soluble fraction in supernatant was collected by centrifuging at 13200 rpm 30 min. Total 1 ml supernatant, which equal to protein expression in 1ml BL21(DE3), was used in SDS-PAGE and Coomassie Blue staining. To calculate the protein expression at different time points, we quantity the image intensity of LbCas12a protein and BSA standard by Image J.
- Coomassie blue staining shows the LbCas12a protein expression was absent at 0 hr and increased as time goes on. This result indicated that T7 promoter can be activated by IPTG-induced T7 polymerase (Figure 1).
- We then used BSA standard to quantify the LbCas12a protein expression. The simple linear regression of BSA protein expression standard is shown in Figure 2, while the original image intensity detected by Image J is shown in table 1.
- Finally, we quantify the LbCas12 expression by conversing the intensity to concentration through BSA standard regression (Figure 3).
| BSA (ng) | Background intensity | BSA intensity | BSA - Background intensity |
| 350 | 2407241 | 1258862 | 1148379 |
| 210 | 1589271 | 962468 | 626803 |
| 70 | 1544912 | 904623 | 180999 |
| 35 | 1042670 | 861671 | 129877 |
| Time (hours) | Background intensity | LbCas12a intensity | LbCas12a - Background intensity | LbCas12a (ng) | LbCas12a (µg/mL) |
| 0hr +IPTG | 696645 | 691258 | 5387 | 8.937 | 2.554 |
| 4hr +IPTG | 900536 | 760105 | 140431 | 50.098 | 14.314 |
| 8hr +IPTG | 925618 | 789444 | 136174 | 48.801 | 13.943 |
| 10hr +IPTG | 913959 | 791540 | 122419 | 44.608 | 12.745 |
| 12hr +IPTG | 956220 | 770282 | 185938 | 63.968 | 18.277 |
| 14hr +IPTG | 976568 | 761481 | 215087 | 72.853 | 20.815 |
| 16hr +IPTG | 1064337 | 764543 | 299794 | 98.671 | 28.192 |
| 16hr Ctrl | 924162 | 743361 | 180801 | 62.403 | 17.829 |
Conclusion:
The quantification of LbCas12a protein expression by BSA standard shows that the Cas12 protein is increased at different time points, which confirmed that the T7 promoter is functional in our construct.
iGEM Marburg 2021 - Contribution
Cell-Free systems are in-vitro tools that are used as prototyping platforms for metabolic networks and genetic constructs. Typically, they are based on crude cell extracts and contain the whole machinery needed for protein biosynthesis. These advances in cell-free systems offer exciting opportunities to fundamentally transform synthetic biology. It enables new approaches for model-driven design of synthetic gene networks, rapid and portable acquisition of targeted components, on-demand biomanufacturing, and building cells from scratch.
In our project we have developed cell-free systems of chloroplasts of various plants. With our Chloroplast cell-free system we provide a testbed that can allow others to test various genetic constructs that will be needed for plant engineering in a shorter time frame.
The DNA concentration response of the T7 polymerase in a chloroplast cell-free system
A significant difference in expression in cell-free systems is caused by the addition of different DNA concentrations[1]. Therefore we wanted to test the behavior of our system to different DNA concentrations.
In the following experiment, the Nanoluc luciferase was used as a reporter system, within our cell-free expression measurement. The graph indirectly shows the expression levels of this Nanoluc luciferase via the emitted luminescence. Extracts from two different species have been utilized and DNA concentrations in the spectrum from 0.5nM to 15nM have been added to the final cell-free reaction to analyse the effect of the DNA concentration on the expression level.
For both N. tabacum and S. oleracea, an optimum is reached at a DNA concentration of 5nM. With a further increase of the concentration, no significant difference in the luminescence can be recognised.
These results show that it is of great importance for experiments using cell-free systems to carefully normalize the DNA concentration for the comparison of a variety of parts.
Additionally, using saturated DNA concentrations has the advantage of being less prone to variations in expression, caused by the use of different concentrations in DNA.
Moreover it should be “good practice” to not have DNA concentrations as a limiting factor in your measurements, as this could cover other effects/properties one would like to actually characterize in the experiment.
References
[1] Kopniczky, M. B., Canavan, C., McClymont, D. W., Crone, M. A., Suckling, L., Goetzmann, B., Siciliano, V., MacDonald, J. T., Jensen, K., & Freemont, P. S. (2020). Cell-Free Protein Synthesis as a Prototyping Platform for Mammalian Synthetic Biology. In ACS Synthetic Biology (Vol. 9, Issue 1, pp. 144–156). American Chemical Society (ACS). https://doi.org/10.1021/acssynbio.9b00437
//chassis/prokaryote/ecoli
//direction/forward
//plasmid/expression/t7
//promoter
//regulation/constitutive
//rnap/bacteriophage/T7
| None |