Part:BBa_K3168002
LargeBitNanoLuc
This basic part codes for the large bit of NanoLuc, which can be used as a split reporter. In combination with the small bit of NanoLuc and the addition of furimazine (the substrate), bright blue light is emitted (Figure 1). A flexible (GGS)5 linker is located in front of the large bit to enable the formation of fusion proteins. A strep-tag is also included at the end for protein purification.
Figure 1. Split-NanoLuc principle.
Usage and Biology
Split reporters have been used a lot to detect protein-protein interactions and for screening purposes (Dixon, 2015). NanoLuc is a small, structurally robust and very bright luciferase. Many methods have been developed using the split variant of NanoLuc, such as the HiBiT technology and NanoLuc ternary technology (Ohmuro-Matsuyama, 2019).
2020 Improvement UZurich
For our 2020 iGEM project, we from the University of Zürich designed a novel approach for a biosensing system relying on plant pattern-recognition receptors (PRRs). We managed to engineer parts of the system in the lab and conduct initial experiments to characterize our system. After many discussions with experts, we decided to use the split-NanoLuc system to generate a luminescent output. Since our system was designed for our chassis S. cerevisiae we decided to use a codon optimized sequence for the output, as we wanted to facilitate expression in our chassis. For this purpose, we codon optimized this part and also Part:BBa K3168003, which encodes for the SmallBit, the second half of the functional NanoLuc luciferase. The registry numbers for the codon optimized parts are : BBa_K3610014 for LargeBit and BBa_K3610014 for SmallBit. As we were using these parts in our project anyway, we decided to test our improved parts and compare them with the original sequences.
Approach
Our goal was it to test whether expression of the codon optimized parts in S. cerevisiae would enhance luminescence intensity of a sample when the NanoLuc substrate furimazine is added.
We tackled this question by using rapamycin and its binding ligands, FKBP and FRB (Part:BBa_K3610022, Part:BBa_K3610023).
FKBP
he FK506 binding protein, or FKBP, is a protein belonging to the immunophilin family and has proven to be a useful tool in biological research. Functionally, the protein is a peptidyl-prolyl cis-trans isomerase (PPI).
Responsible for its prominent role in research is the ability of FKBP to bind to rapamycin, an antifungal antibiotic macrolide. What makes this interaction interesting is the fact that rapamycin binds to FKBP and the FKBP–rapamycin binding (FRB) domain of the mammalian target of rapamycin (mTOR) simultaneously, inducing a dimerization of these two components. These properties of FKBP and FRB have been exploited for conditional dimerization of proteins of interest, which offers many possibilities to artificially manipulate cellular processes, by fusing them to FKBP and FRB.
FRB
The mammalian target of rapamycin (mTOR) is a Ser/Thr protein kinase, which is involved in the control of mRNA translation initiation. The FKBP12-rapamycin binding domain (FRB) is located upstream of the catalytic domain and is essential for the kinase activity of the mTOR protein. Without the FRB domain the protein loses its functionality. As already established the interesting property of FRB is its interaction with the antifungal antibiotic macrolide rapamycin. Rapamycin binds to FRB and the FK506 binding protein (FKBP) simultaneously, inducing dimerization of the two components. This rapamycin-induced dimerization has been exploited for dimerization of proteins of interest by fusing them to FKBP and FRB.
Rapamycind binding ligands and NanoBits
The previously described properties come in handy when we need to induce protein-protein interaction. We decided to test the improved construct in a dimerization assay with rapamycin.
For this purpose, we designed the following constructs:
- Part:BBa_K3610054 LargeBit fused to FKBP12 (LBit:FKBP), Part:BBa_K3610054 optimized LargeBit fused to FKBP12 (LBit:FKBP_yeast), Part:BBa_K3610054 SmallBit fused to FRB (SBit:FRB), Part:BBa_K3610054 optimized SmallBit fused to FRB (SBit:FRB yeast).
The split protein was fused to either FKBP12 or FRB via a 15 amino acid(AA)-long linker consisting of the small AAs Glycin and Serine. Additionally, the two LargeBit constructs contain a hemagglutinin tag at between the receptor protein-coding region and the 15AA-linker, which is used for (co-)immunoprecipitation and Western Blotting. The SmallBit constructs have a FLAG tag at the same position for the same purpose.
For testing our newly created part Part:BBa_K3610054, we co-expressed LBit:FKBP and SBit:FRB in <S. cerevisiae (AP4)</i> and did the same for LBit:FKBP_yeast and SBit:FRB. When both parts are expressed, addition of rapamycin will make the ligands FKBP12 and FRB bind to it, which will initiate the interaction between the LargeBit and the SmallBit. Since SmallBit is very short and has hardly any codons that are rare in S. cerevisiae, we only tested the codon optimized LargeBit. Should codon optimization actually facilitate expression in yeast, we would expect the luminescence intensity to be higher, when rapamycin and the substrate furimazine are added.
Preparation of the constructs
In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed E. coli (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same E. coli strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI.
We expressed the S. cerevisiae cells with two different plasmids, each containing one construct. One plasmid with a -TRP gene, enabling the yeast strain to produce tryptophan, the other contains a gene for the kanamycin acetyltransferase. We therefore needed a synthetic dropout medium (-TRP) supplemented by Geneticin.
Procedures
After successful transformation, we examined the cells with a plate reader of the type Synergy H1. Cells were inoculated in liquid medium for several hours at 30°C and adjusted to the same OD600. This step was necessary to prevent differences in measured values due to higher cell density in one sample.
Afterwards, the 96 well plate was prepared with 50μl samples containing LBit and samples containing LBit_yeast. We divided the samples into groups, those to which no rapamycin was added and those that received additional rapamycin. The substrate furimazine was added to each well in form of 50μl NanoGlo solution (50:1 buffer to substrate). The rapamycin was added immediately after addition of furimazine. 10μM rapamycin was added, giving a final concentration of 10 μM.
Further Details:
- Runtime 2:00:00 (HH:MM:SS)
- Interval 0:06:00, 21 Reads
- Shake
- Linear:
- Continuous
- Frequency: 493 cpm (4 mm)
- Read
- Luminescence Endpoint
- Full Plate
- Integration Time: 0:03.00 (MM:SS.ss)
- Filter Set 1
- Emission: Full light
- Optics: Top, Gain: 135
- Read Speed: Normal, Delay: 100 msec
- Extended Dynamic Range
- Read Height: 1 mm
- Linear:
Results
Assay 1
In the first measurement series, all samples were adjusted to OD600 = 0.26. The following samples were prepared:
- 6X LBit_yeast without rapamycin
- 6X LBit yeast with rapamycin
- 6X LBit registry without rapamycin
- 6X LBit registry with rapamycin
(LBit registry referres to this part)
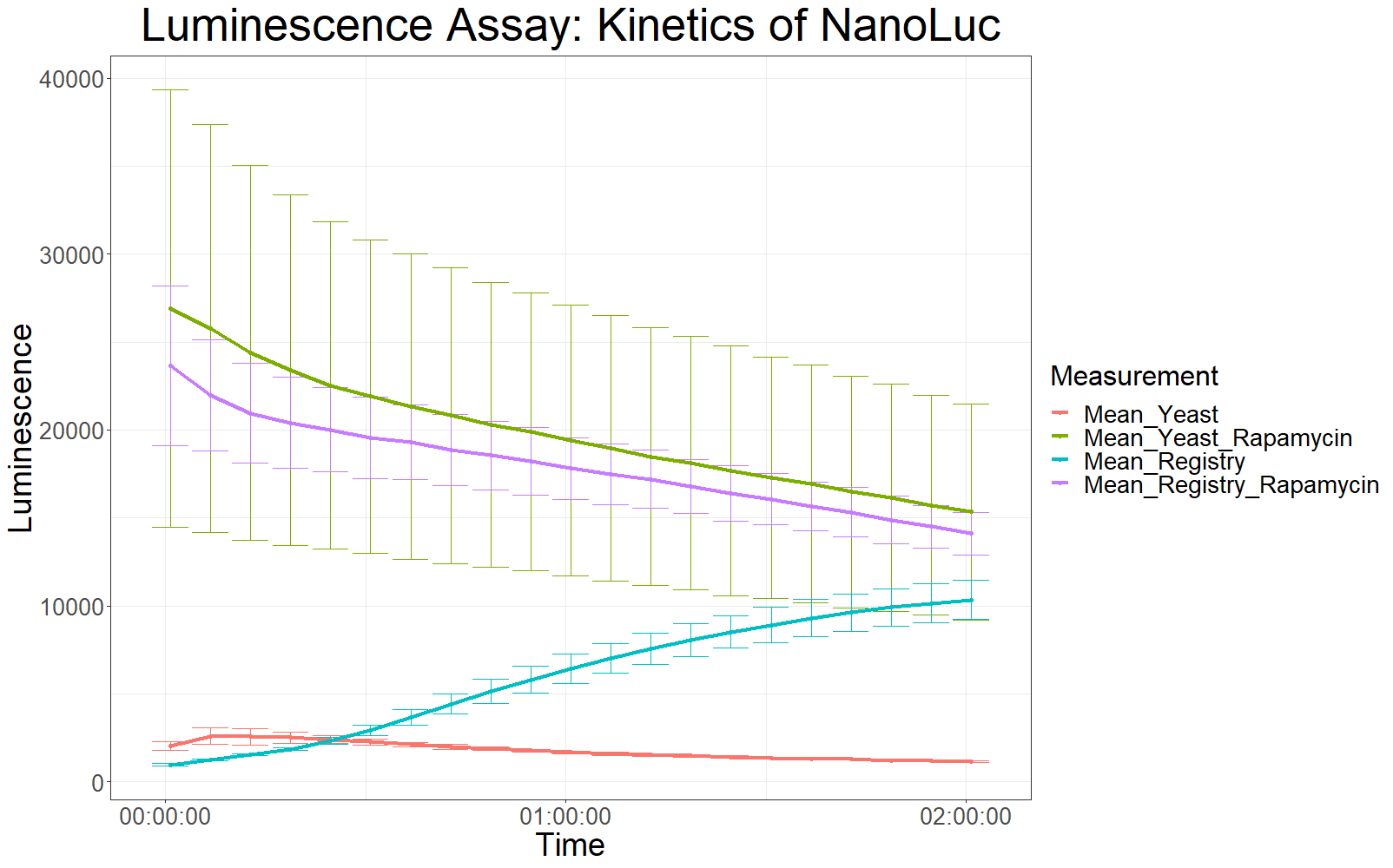
Measurements with a luminometer at 480 nm over two hours gave the following results.
| Time | Yeast | Yeast | Yeast | Yeast | Yeast | Yeast | Yeast + rapamycin | Yeast + rapamycin | yeast + rapamycin | yeast + rapamycin | yeast + rapamycin | yeast + rapamycin | registry | registry | registry | registry | registry | registry | registry + rapamycin | registry + rapamycin | registry + rapamycin | registry + rapamycin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00:00:45 | 2430 | 2106 | 2092 | 1994 | 1895 | 1710 | 49039 | 32120 | 13199 | 20462 | 22483 | 24117 | 1042 | 970 | 954 | 1018 | 877 | 903 | 20088 | 20125 | 29619 | 24759 |
| 00:06:45 | 3271 | 2816 | 2911 | 2204 | 2315 | 2026 | 46768 | 30463 | 13767 | 19967 | 21892 | 21831 | 1310 | 1244 | 1272 | 1294 | 1217 | 1212 | 20065 | 19926 | 26624 | 21277 |
| 00:12:45 | 3214 | 2820 | 2889 | 2178 | 2244 | 2056 | 43677 | 28730 | 13313 | 18920 | 20964 | 20778 | 1550 | 1545 | 1580 | 1569 | 1493 | 1479 | 19268 | 19546 | 25222 | 19791 |
| 00:18:45 | 3023 | 2650 | 2664 | 2287 | 2305 | 2100 | 41396 | 27459 | 12911 | 18189 | 20333 | 20162 | 1812 | 1921 | 1997 | 1826 | 1806 | 1792 | 18789 | 19224 | 24308 | 19327 |
| 00:24:45 | 2763 | 2467 | 2459 | 2330 | 2314 | 2070 | 39354 | 26259 | 12778 | 17623 | 19618 | 19525 | 2185 | 2420 | 2639 | 2278 | 2202 | 2252 | 18502 | 19078 | 23598 | 18899 |
| 00:30:45 | 2552 | 2270 | 2301 | 2248 | 2260 | 2021 | 38112 | 25162 | 12574 | 17283 | 19360 | 18904 | 2629 | 3095 | 3486 | 2799 | 2771 | 2797 | 17938 | 18721 | 22991 | 18565 |
| 00:36:45 | 2365 | 2085 | 2082 | 2094 | 2104 | 1941 | 37227 | 24339 | 12385 | 16842 | 18745 | 18397 | 3188 | 3925 | 4386 | 3451 | 3456 | 3517 | 17790 | 18598 | 22435 | 18363 |
| 00:42:45 | 2234 | 1939 | 1928 | 1979 | 2006 | 1849 | 36265 | 23546 | 12096 | 16723 | 18341 | 17976 | 3827 | 4780 | 5369 | 4094 | 4136 | 4256 | 17356 | 18112 | 21884 | 18116 |
| 00:48:45 | 2090 | 1850 | 1851 | 1857 | 1884 | 1751 | 35178 | 22867 | 11895 | 16327 | 17988 | 17540 | 4400 | 5586 | 6271 | 4822 | 4857 | 4929 | 16951 | 17857 | 21400 | 17986 |
| 00:54:45 | 1923 | 1736 | 1720 | 1747 | 1801 | 1655 | 34509 | 22317 | 11848 | 16050 | 17505 | 17175 | 4890 | 6314 | 7066 | 5422 | 5473 | 5622 | 16600 | 17608 | 21021 | 17674 |
| 01:00:45 | 1812 | 1652 | 1609 | 1676 | 1705 | 1586 | 33544 | 21861 | 11442 | 15757 | 17115 | 16677 | 5431 | 7007 | 7811 | 6030 | 6111 | 6203 | 16252 | 17150 | 20335 | 17502 |
| 01:06:45 | 1756 | 1549 | 1505 | 1603 | 1626 | 1484 | 32964 | 21117 | 11353 | 15354 | 16719 | 16285 | 5945 | 7511 | 8433 | 6639 | 6739 | 6801 | 15953 | 16864 | 19968 | 17190 |
| 01:12:45 | 1670 | 1503 | 1495 | 1515 | 1551 | 1468 | 32209 | 20543 | 11228 | 14970 | 16261 | 15748 | 6463 | 8079 | 9036 | 7137 | 7257 | 7326 | 15692 | 16609 | 19538 | 16976 |
| 01:18:45 | 1592 | 1438 | 1430 | 1468 | 1485 | 1383 | 31574 | 20053 | 10939 | 14737 | 15890 | 15547 | 6883 | 8649 | 9555 | 7528 | 7735 | 7883 | 15338 | 16182 | 18899 | 16729 |
| 01:24:45 | 1526 | 1353 | 1366 | 1409 | 1441 | 1347 | 31014 | 19482 | 10631 | 14317 | 15546 | 15120 | 7310 | 9101 | 9996 | 8100 | 8217 | 8390 | 14870 | 15861 | 18607 | 16267 |
| 01:30:45 | 1444 | 1309 | 1321 | 1357 | 1372 | 1297 | 30195 | 18987 | 10583 | 14070 | 15144 | 14679 | 7581 | 9525 | 10558 | 8470 | 8582 | 8792 | 14594 | 15499 | 18019 | 16121 |
| 01:36:45 | 1420 | 1271 | 1277 | 1281 | 1305 | 1241 | 29662 | 18598 | 10246 | 13906 | 14769 | 14456 | 7899 | 9965 | 10959 | 8914 | 8873 | 9234 | 14334 | 15107 | 17597 | 15632 |
| 01:42:45 | 1360 | 1266 | 1249 | 1292 | 1278 | 1217 | 28843 | 18010 | 10022 | 13499 | 14429 | 14021 | 8216 | 10295 | 11263 | 9165 | 9292 | 9450 | 13909 | 14814 | 17252 | 15296 |
| 01:48:45 | 1284 | 1183 | 1183 | 1181 | 1241 | 1197 | 28291 | 17679 | 9855 | 13161 | 14198 | 13638 | 8449 | 10496 | 11649 | 9449 | 9695 | 9743 | 13500 | 14327 | 16701 | 15007 |
| 01:54:45 | 1266 | 1178 | 1139 | 1189 | 1238 | 1105 | 27502 | 17233 | 9583 | 12973 | 13746 | 13337 | 8616 | 10862 | 11888 | 9623 | 9843 | 10037 | 13216 | 14020 | 16099 | 14695 |
| 02:00:45 | 1218 | 1110 | 1131 | 1123 | 1182 | 1151 | 26890 | 16840 | 9330 | 12579 | 13572 | 12858 | 8885 | 10994 | 12129 | 9834 | 10045 | 10175 | 12851 | 13614 | 15693 | 14332 |
The measurement values are summarized in the graph below:
As expected, the control samples (no rapamycin added) gave hardly any detectable luminescent output. When rapamycin was added to the wells, increased luminescence intensity could be detected. This initial assay showed increased luminescence in samples transformed with LBit:FKBP_yeast (codon optimized). The variance, however, is too big to make any assumptions after this initial measurement (regarding increased luminescence when compared to the original sequence).
Assay 2
Because the first data from the first measurement was not very conclusive, the same type of measurement was repeated. With the following differences:
- OD600 = 0.5 for all samples
- longer waiting period between taking cells out of the incubator and measurement (2 hours)
- After addition of NanoGlo solution, the samples were incubated at RT for 30 minutes.
The following samples were measured:
- 3X yeast without rapamycin
- 9X yeast with rapamycin
- 3X registry without rapamycin
- 3X registry with rapamycin
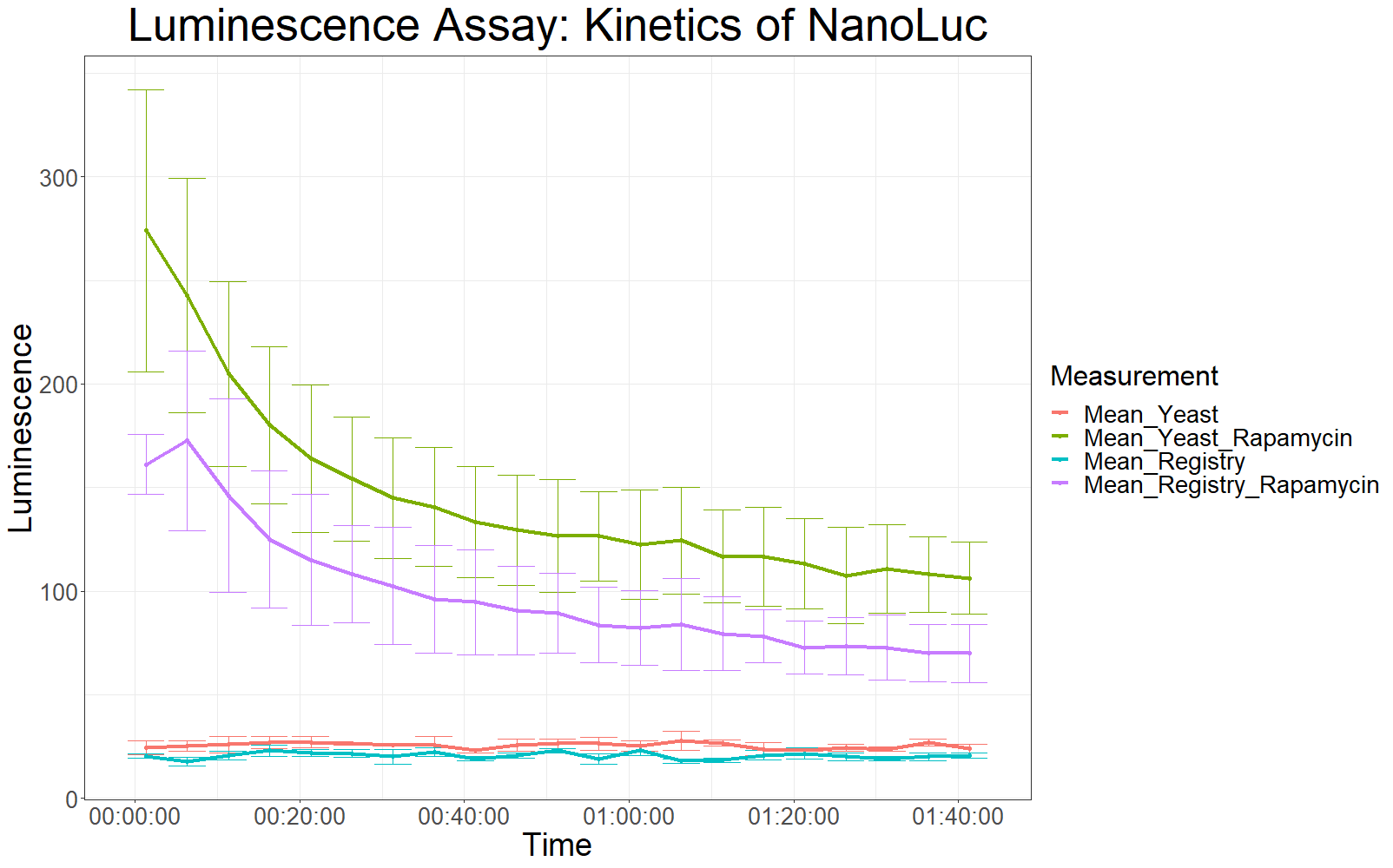
The patterns observed in the second experiment were similar to the ones observed in the first assay. Expression of the constructs was successful and rapamycin was able to drive the interaction between the LargeBit and the SmallBit part, which is made visible by the luminescent output after addition of the NanoLuc substrate furimazine.
As had already been previously observed, samples which expressed the codon optimized version of this part, showed higher average fluorescence intensity at all measurement points.
Conclusion
We conducted initial tests to characterize the parts Part:BBa K3168002 and Part:BBa_K3610014. When SmallBit and LargeBit are fused to the two rapamycin binding ligands FRB and FKBP12 respectively and expressed in S. cerevisiae (AP4), addition of rapamycin is able to drive the interaction between the SmallBit and the LargeBit, leading to the reconstitution of their functionality as a luciferase. Addition of rapamycin and the substrate furimazine will lead to a luminescent output.
Additionally, initial measurements with a luminometer suggested a stronger overall luminescence intensity, when the codon optimized sequence of LargeBit was used.
References
Dixon, A. S., Schwinn, M. K., Hall, M. P., Zimmerman, K., Otto, P., Lubben, T. H., ... & Wood, M. G. (2015). NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS chemical biology, 11(2), 400-408.
Ohmuro-Matsuyama, Y., & Ueda, H. (2019). Protein-Protein Interaction Assays Using Split-NanoLuc. In Bioluminescence. IntechOpen.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 526
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |