Part:BBa_K3380500
iSpinach fluorescent RNA aptamer construct under T7 RNA polymerase promoter (BBa_z0251)
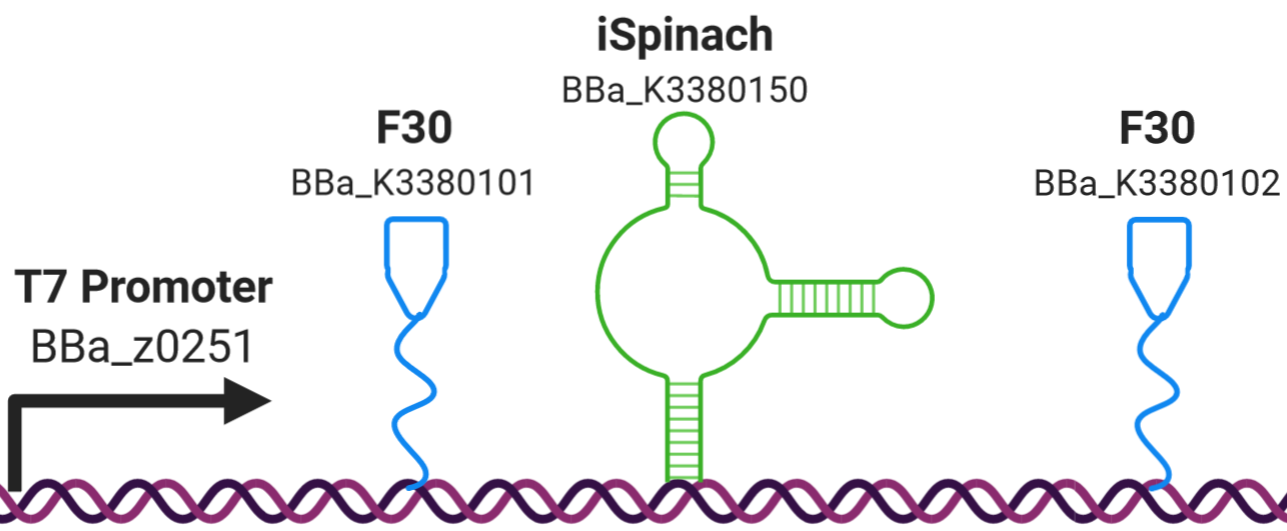
The Edinburgh iGEM team 2020 designed a construct comprising a fluorescent RNA aptamer (iSpinach BBa_K3380150) flanked by a tRNA scaffold (F30 BBa_K3380101 and BBa_K3380102) under a strong class III T7 RNA polymerase promoter (BBa_z0251). It was designed to test the fluorescence intensity of the iSpinach aptamer flanked by F30 tRNA scaffold when binding to the DFHBI fluorophore. The construct is capable of exhibiting fluorescence being a transcription only construct. Simultaneously we tested the efficiency of transcription in the absence of a terminator. The T7 RNA polymerase is capable of synthesizing transcripts via run-off transcription in the absence of a terminator. Figure 1 illustrates a schematic design of the construct.
Figure 1: Construct BBa_K3380500 design. The iSpinach fluorescent RNA aptamer (shown in green) was flanked by the F30 upstream and downstream scaffolds (shown in blue) to protect it from the RNAse degradation (from the cell free extract), it was expressed under the Class III strong T7 RNA polymerase promoter BBA_z0251 (shown in black).
Usage and Biology
It can be used in cell-free extracts or buffers containing T7 RNA polymerase, chemical energy (ATP), NTPs, fluorophore and adequate cofactors and pH to test the fluorescence intensity of the iSpinach aptamer (BBa_K3380150) when bound to fluorophores under the T7 promoter. It might exhibit lower fluorescence, due to degradation by the RNAses, depending on the cell extract it is used in. However, the absence of the scaffold can also improve the fluorescence, as in the example of Broccoli fluorescent RNA aptamer.
The construct functioning solely on transcription and having a small length of only 195 nucleotides has multiple advantages over the conventional fluorescent proteins. The time required for the fluorescence formation is much shorter than compared to the time needed for the synthesis of the fluorescent proteins. Moreover, there is less burden exhibited on the cell (if expressed in cells) or it requires less resources (NTPs, energy if expressed in cell free extract) when expressing the transcription only construct. Also, individual parts can be "de novo" synthesized and ligated rather than expressing them in cells using plasmids, which is more expensive and requires more time.
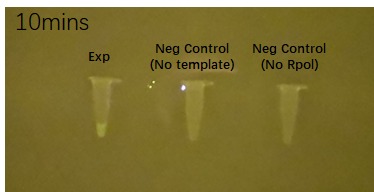
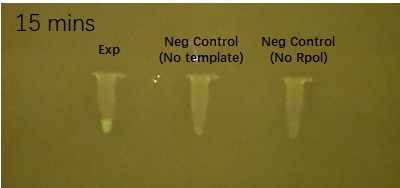
| DFHBI fuorophore binding to the BBa_K3380500 construct | ||
|---|---|---|
| 5 minutes after fluorophore addition | 10 minutes after fluorophore addition | 15 min after fluorophore addition |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 25
| None |