Part:BBa_K863010
tthl laccase from Thermus thermophilus with T7 promoter, RBS and His-tag
tthl (Laccase from Thermus thermophilus) with T7, RBS and HIS tag
Usage and Biology
Slovenia HS characterized this part in 2015.
Escherichia coli BL21 (DE3) bacteria were transformed with the expression plasmids (BBa_K863005 and BBa_K863010) and grown in 10 ml at 37 °C in LBC medium overnight. To express both recombinant proteins, 10 ml of overnight cultures shaker cultures were grown at 37 °C in LB broth supplemented with 30 µg/ml chloramphenicol and shaking with 225 rpm. Expression of the recombinant protein was induced by addition of IPTG to a final concentration of 1 mM, when the cell density reached an OD600 of 0.8. After induction, cells were grown for 5 h and then collected by centrifugation at 6000g for 10 min. The cell pellet collected from 400 ml of bacterial culture was resuspended in 20 ml of resuspension buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole) and sonified 3 × 6 min on ice. Following centrifugation at 30 000 × g for 10 min to remove insoluble debris, the supernatant was applied to a Ni-NTA Superflow Cartridge (Qiagen) connected to ÄKTA FPLC system, washed with the resuspension buffer and eluted in the same buffer, but containing 250 mM imidazole. The peak fractions were collected and 15 µl of each fraction was collected and resolved on 12 % SDS-PAGE.


We found that while BBa_K863005 shows excellent activity, BBa_K863010 shows no activity under the same conditions.
IPTG Induction of K863010 (2019 PuiChing_Macau)

TecCEM Characterization,
colorimetric assay and purification
For
the characterization of the Laccase BBa_K863010 we conducted an IPTG induction
experiment in which we used the transformation of the Laccase in E. coli
BL21. We thought that we could use another strain called SoluBL21
but results were not successful as no expression was found. We verified the
presence of the protein through an SDS-PAGE with a gel concentration of 12% and
found a visible band with a mass of around 50 kDa. This can be seen in figure
1.

Figure 1. The lanes correspond to
the following. M: Molecular weight marker; 1: Total protein after induction; 2:
Total protein before induction; 3: Protein found in the Culture Medium after
induction; 4: Cytoplasmic soluble fraction; 5: Inclusion bodies of the
insoluble fraction; 6: Concentrated Culture Medium after induction. The band
observed in lanes 1, 3 and 6 weighs around 48 kDa and
corresponds to the expected size.
We
found that the protein was mostly found on the culture medium but can also be
found on the cytoplasmic soluble fraction. The band that was appreciated in
figure 1. indicates that there’s an expression of the Laccase after it’s
induction with IPTG so our results and experience using this part was different
from what 2019 PuiChing Macau’s team reported
previously.
To
prove that the Laccase was being expressed, we conducted a colorimetric assay
involving three colorants that act as a substrate: methylene blue, malachite
green and rose bengal. We based this experiment upon
the findings of D. Singh et al. (2014) [1], in which they used agar plates with
these colorants to determine the expression of Laccases in a medium. The first
assays we conducted were only to find out if there was any color change with
the presence of our extracted Laccase either from the cytoplasmic soluble
fraction or from the culture medium. We used Citrate Buffer and found a change
of color in different samples of Laccase after its purification using Ni
Affinity. We also verified that the effect we saw wasn’t related to a change in
pH. The results
are available in figure 2.

Figure 2. In section a) we can see
the change of color of the substrates used (methylene blue, malachite green and
rose bengal from left to right). a)-I. corresponds to
the cytoplasm soluble fraction while a)-II. corresponds to the secreted protein
from the culture medium. In section b) we can see that the pH remained
unchanged throughout the assay.
After
this first assay, we decided that we had to establish a purification protocol
through which we could get the most enzyme possible. We used a Ni Affinity Column
and a system of recollection of the different phases. We collected the enzyme
from both the culture medium and the cytoplasmic soluble fraction and measured
the absorbance of the fractions collected at 280 nm. In total, we got 21
fractions for the cytoplasm proteins and 20 fractions for the culture medium.
The purification conditions were established using a growing elution buffer
concentration. These conditions
are shown in figures 3 and 4.

Figure 3. Chromatogram of the
purification of the cytoplasmic soluble fraction showing the spike of
absorbance in an elution volume of around 15 mL to 20 mL (with a percentage of
elution buffer of around 30 to 50%); corresponding to the fractions containing
the Laccase.
>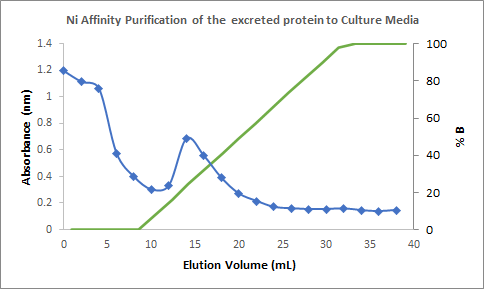
Figure 4. Chromatogram of the
purification of the culture medium showing the spike of absorbance in an
elution volume of 12 mL to 20 mL (with a percentage of elution buffer of around
16% to 50%); corresponding to the fractions containing the Laccase.
Finally,
we conducted a last assay in which we first quantified the amount of protein
recovered through a BCA quantification protocol. Using this protocol and with
the elaboration of a BCA curve, we estimated that we recovered 0.334 µg/mL of
Laccase in the culture medium while for the cytoplasmic soluble fraction we
obtained 0.1298 µg/mL of Laccase. This was consistent with the results we got
from the chromatograms.
For
the final colorimetric assay we conducted, we quantified the activity of the
Laccase obtained from the purification. We compared it with a Commercial
Laccase from Sigma belonging to Trametes
versicolor and used the spectrophotometer to measure methylene blue,
malachite green and rose bengal at 664, 617 and 562
nm respectively. Since we didn’t quite have the exact concentration of
colorants in our samples, we limited to measure the activity as a percentage of
degradation of each colorant where a 100% of substrate would be the absorbance
of the control of the blue, green and rose colorants
and the enzymatic degradation would be expressed as the loss of color. The results can be seen below.
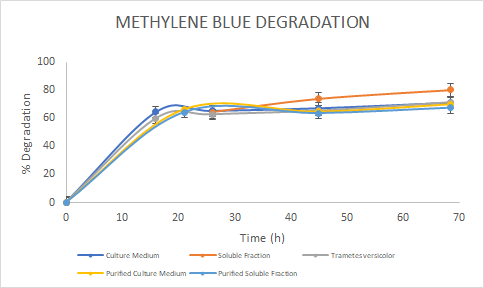
Figure 5. Percentage of
degradation of Methylene Blue through time for the Laccase in the culture
medium (blue), soluble fraction (orange), purified culture medium (yellow),
purified soluble fraction (light blue) and the Laccase from Trametes
versicolor (gray).
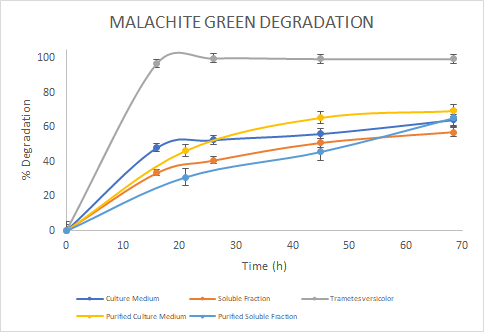
Figure 6. Percentage of
degradation of Malachite green through time for the Laccase in the culture
medium (blue), soluble fraction (orange), purified culture medium (yellow),
purified soluble fraction (light blue) and the Laccase from Trametes
versicolor (gray).
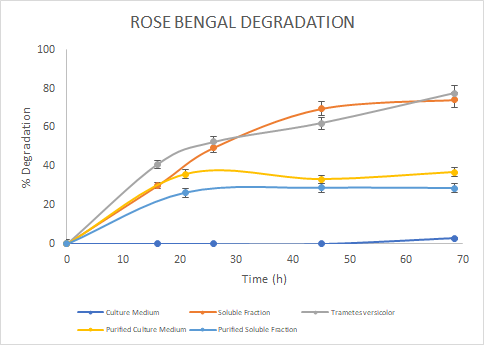
Figure 7. Percentage of
degradation of Rose Bengal through time for the Laccase in the culture medium
(blue), soluble fraction (orange), purified culture medium (yellow), purified
soluble fraction (light blue) and the Laccase from Trametes
versicolor (gray).
In
the assays, we saw a similar trend for the degradation of Methylene Blue. For
Malachite Green, the Laccase from Trametes versicolor
had a higher degradation rate. Finally, for Rose Bengal we observed a similar
trend between Trametes versicolor
Laccase and the Soluble Fraction Laccase (before purification). We then
reported the percentage of degradation of each sample for each colorant at the
final point in time and got the next results:

Figure 8. Percentage of
degradation of each Laccase at the final point in time for each colorant.
We
observed that overall, the best results were obtained by the Laccase of Trametes versicolor (as expected) followed by
the soluble fraction and the purified soluble fraction. However, it is worth
noting that although these results show that the commercial Laccase may have
higher degradation values, it also has a higher concentration since it was
prepared at 1 mg/mL (compared to the purified Laccases which had concentrations
of 0.334 µg/mL in the culture medium and 0.1298 µg/mL in the soluble
fraction.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 1408
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 475
Illegal NgoMIV site found at 962 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 790
//function/degradation
//proteindomain/degradation
| None |
