Part:BBa_K302033
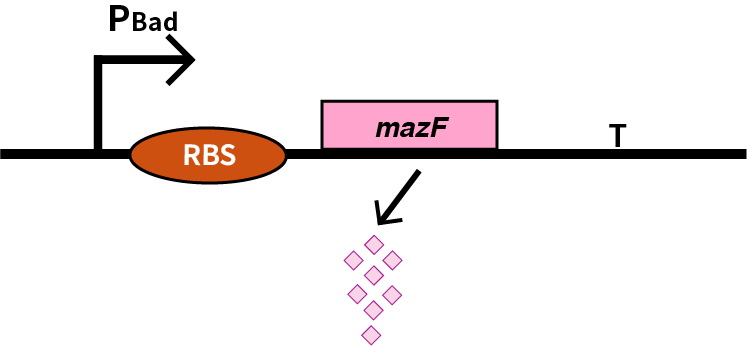
mazF
Encodes a stable non-specific ribonuclease in Bacillus Subtilis. It is used in conjungtion with mazE in Bacillus Subtilis to provide a toxin-antitoxin kill switch in various stressful conditions. When expression of both genes is turned off (as both are under contol of same promoter) mazE will be degraded faster than mazF. There is then no inhibiton of mazF, killing the cell.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterized by BNU-China 2019
We characterize mazF (BBa_K302033) by an induced suicide system, in which the downstream mazF (BBa_K302033) gene encoding toxin MazF is put under the control of L-arabinose induced promoter pBAD (BBa_K206000). MazF is an endoribonuclease that cleaves RNAs at ACA sites and causes the death of microbe as a reporter [1]. As a result, we can characterize mazF in a cell density-dependent manner in Escherichia coli K-12.
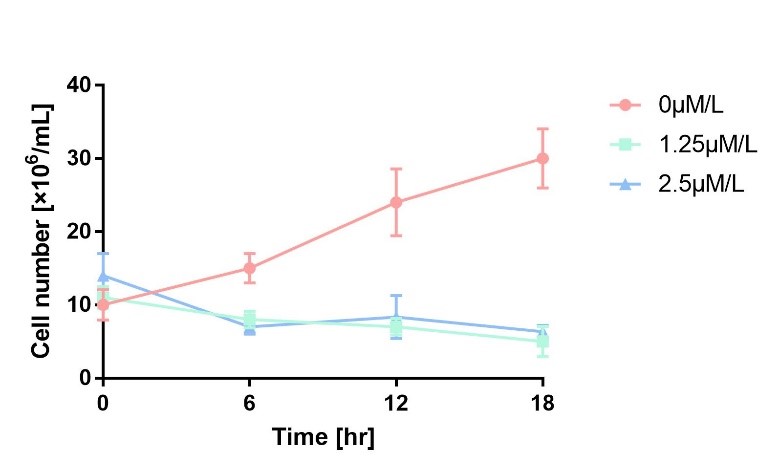
In order to characterize the toxicity of protein encoded by mazF, we take engineered microbe without induction as control group. As is shown in Fig.1, the cell number of experimental groups show a significant decrease upon induction, which indicates the protein encoded by mazF is lethal to E. coli.
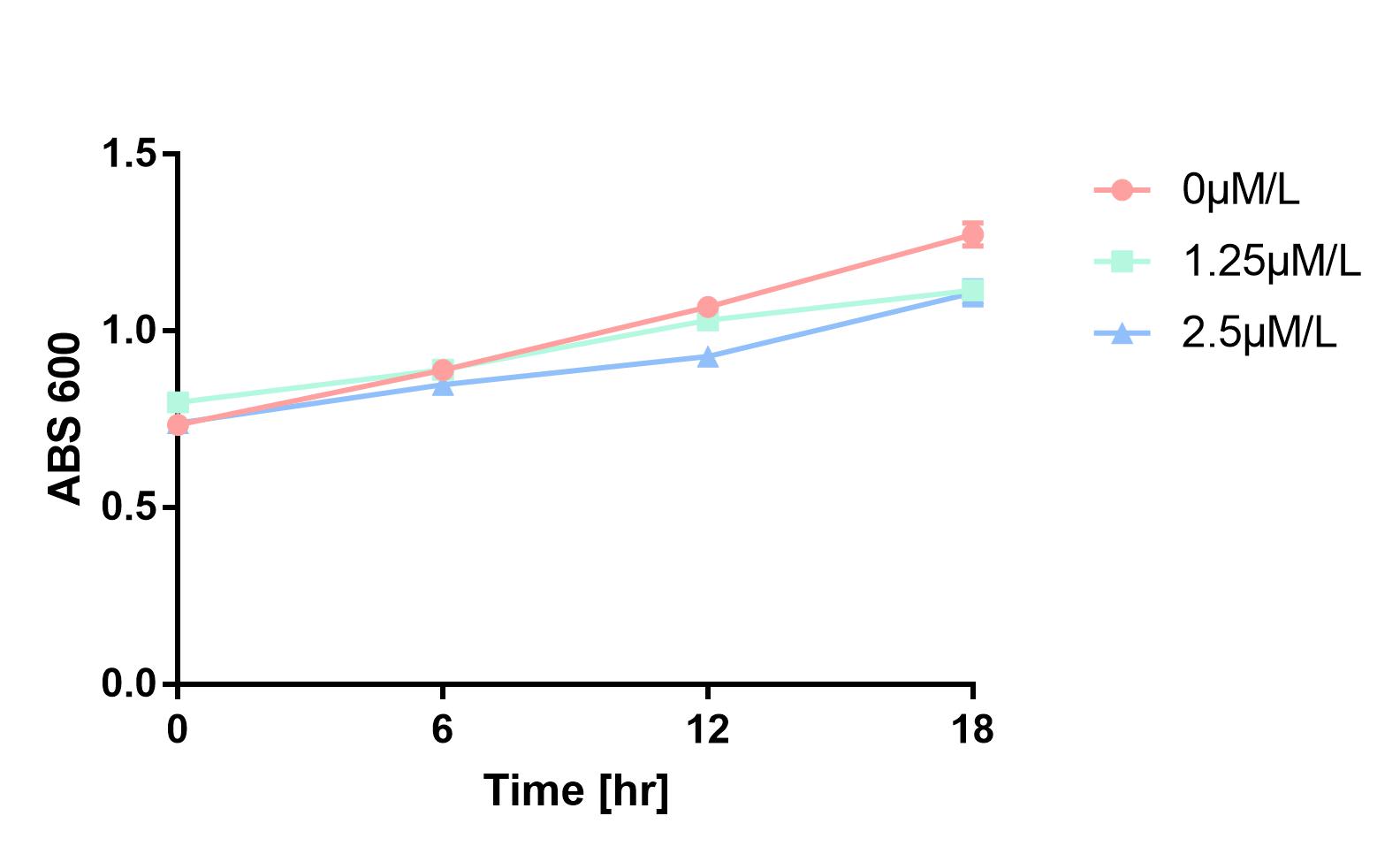
Furthermore, in order to prove that MazF kills the cell without lysing it, we measure OD600 of each sample to give an overall number of intact bacteria, dead and alive. As is shown in Fig.2, there is little difference between control and experimental groups, although it is validated numbers of alive cells differ. Hence, we reach a conclusion that protein encoded by mazF mediates suicide of cells without causing lysis of bacterial cells.
Experimental approach
1. Transform the plasmids into E. coli DH5α competent cells.
2. The engineered bacteria are cultured in 200mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm;
3. Equally divide the culture into 90 centrifuge tubes, which is 1mL respectively. Centrifuge them at 4000rpm for 5 minutes. Discard the liquid.
4. Resuspend 30 tubes of collected bacteria with LB-ampicillin (50 ng/µl) containing 1.25μM/L and 2.5μM/L L-arabinose respectively as experimental groups. Resuspend 30 tubes of bacteria with pure LB-ampicillin (50 ng/µl) medium.
5. Collect 3 tubes of all groups every 6 hours, dilute all of the samples to 10^7 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately. At the same time, refresh the medium to maintain the concentration of L-arabinose.
6. Count the number of colonies in 5 cm^2 per plate after cultured for 24 hours at 37℃
7. Three repicas are tested in each group.
Reference
[1] Diana Széliová, Ján Krahulec, Martin Šafránek, et al. Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains[J]. Journal of Biotechnology, 2016, 236:1-9.
Characterized by XJTU-China 2020
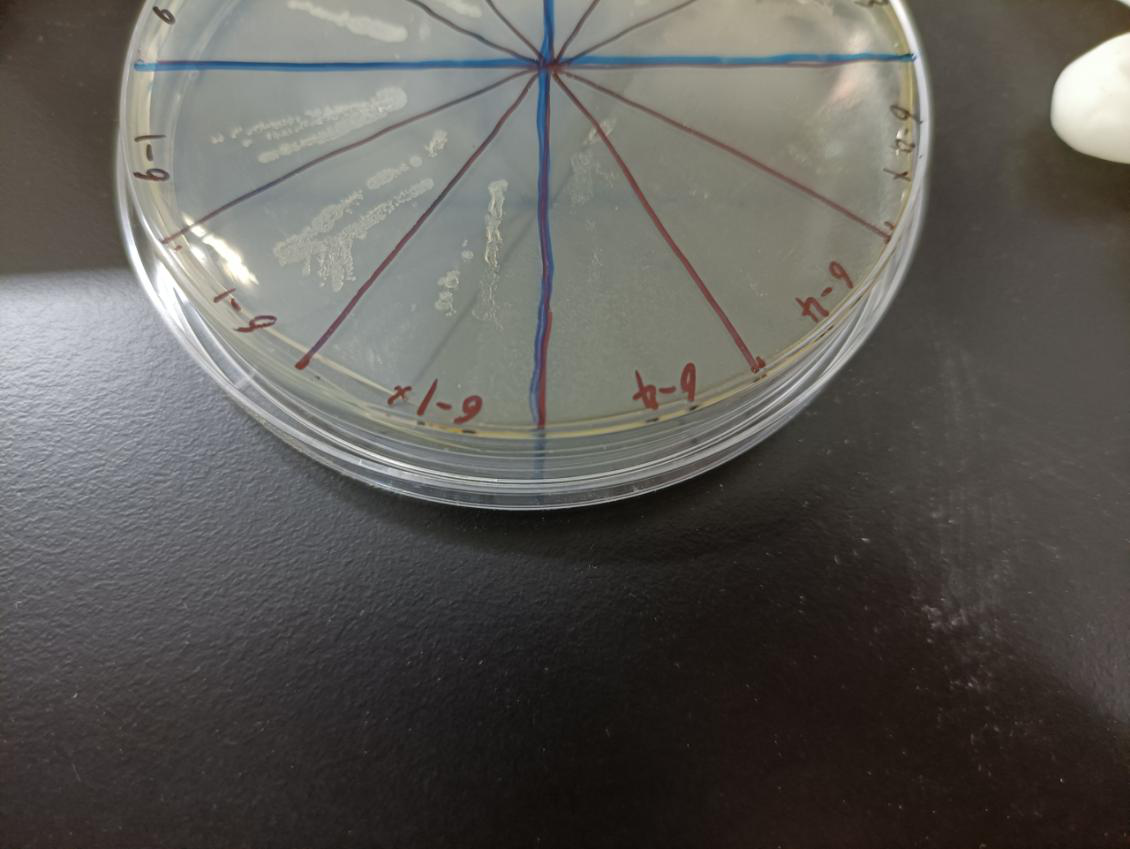
In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 groups were the control group without arabinose induction, and three of 6-4 groups were the experimental group with 1.5 M/L arabinose induction. Where, the dilution factor of the group with * is 10^4, and the dilution factor of the group without * is 10^6.
Result: The bacteria without arabinose induction grow normally, while with arabinose induction, the bacteria suicide.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//chassis/prokaryote/bsubtilis
| None |