Part:BBa_B0035
RBS (B0030 derivative)
-- No description --
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Lambert_GA 2019 Characterization
Lambert_GA 2019 tested several combinations of constitutive promoters and ribosomal binding sites to characterize each by measuring enzyme activity and therefore protein expression. The gene expressed, LacZ, codes for β-galactosidase (β-gal), which typically breaks down lactose. Instead of using lactose, we added the sugar ONPG (Ortho-Nitrophenyl-β-galactoside). β-gal breaks ONPG down into galactose and ONP (Ortho-Nitrophenol), which has a yellow color. If there is more ONP present, there is more enzymatic activity and therefore more expression of LacZ. We used a plate reader to measure absorbance at 420 nm, measuring yellow color, and 600nm, measuring cell density. We inputted those absorbance values into the Miller unit formula to calculate enzymatic activity per cell per milliliter.
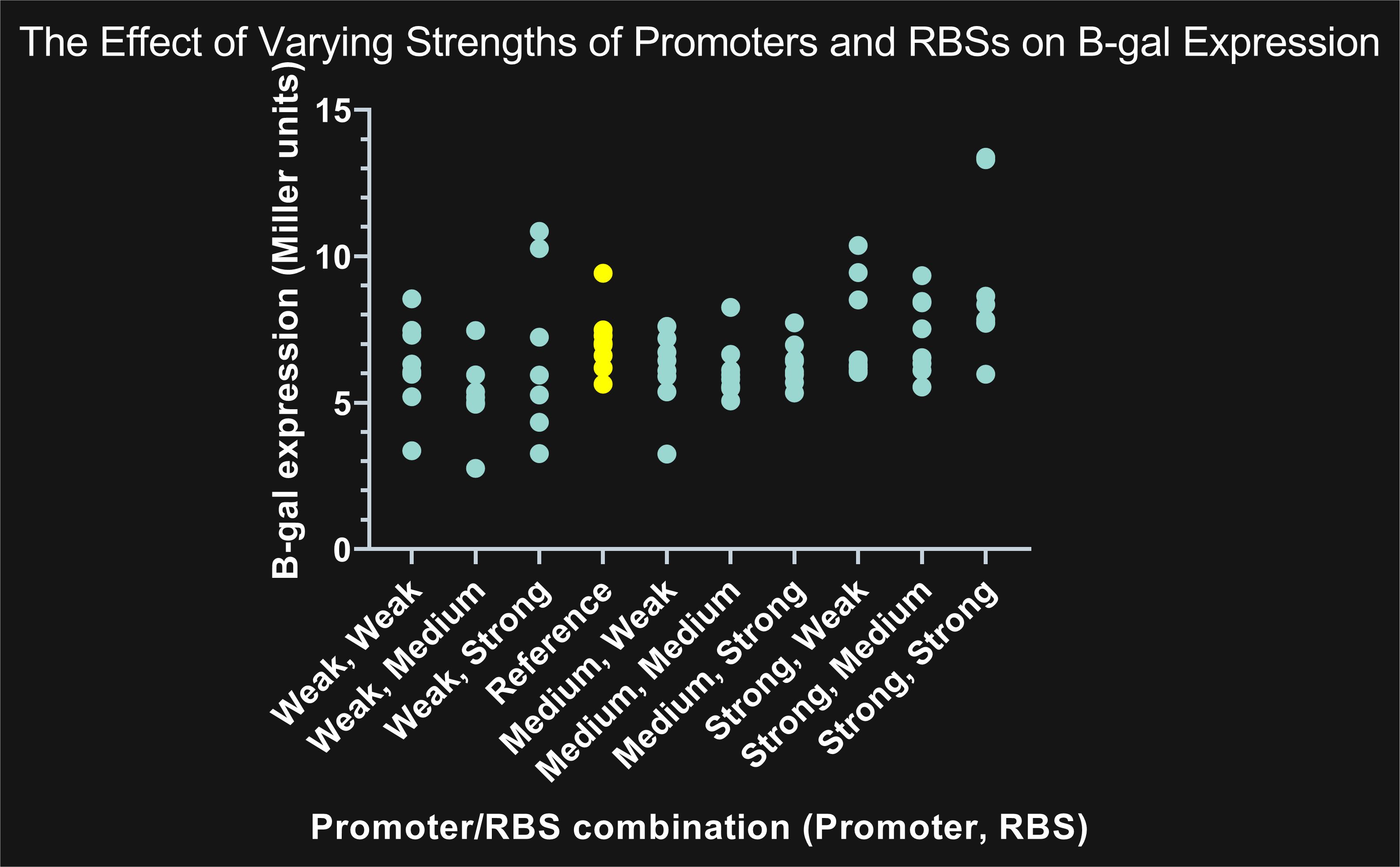 Figure 1: The highlighted points in yellow represent BBa_B0035 (reference RBS) compared to the remaining nine combinations highlighted in blue. Combinations showing promoters with the same strength, but different RBS, share similar expression. An increase in promoter strength results in an increase in expression; on the other hand, changes in RBS strength has a negligible effect on expression.
Figure 1: The highlighted points in yellow represent BBa_B0035 (reference RBS) compared to the remaining nine combinations highlighted in blue. Combinations showing promoters with the same strength, but different RBS, share similar expression. An increase in promoter strength results in an increase in expression; on the other hand, changes in RBS strength has a negligible effect on expression.
//ribosome/prokaryote/ecoli
//chassis/prokaryote/ecoli
//direction/forward
//regulation/constitutive
| None |
