Part:BBa_B0034
RBS (Elowitz 1999) -- defines RBS efficiency
RBS based on Elowitz repressilator.
Usage and Biology
IIT Madras 2016's Characterization
Modelling
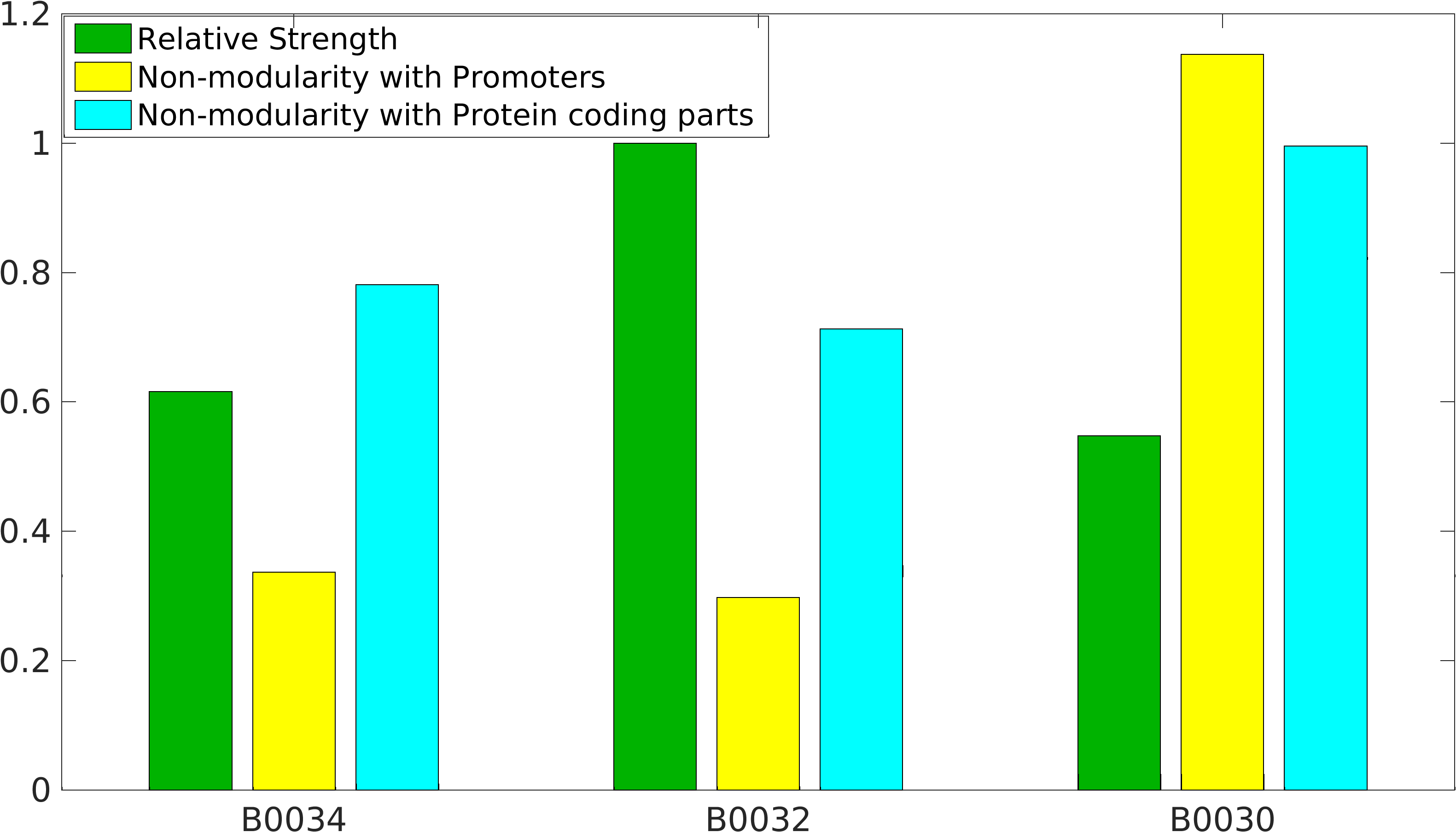
Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]
Experimentation
Biobrick RBSs B0030, B0031, B0032, B0034 were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.
Note: The Elowitz RBS is the definition of efficiency 1.0.
Team Warsaw 2010's measurement
definition of 100% strength in our measurement.Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this RBS.
Documentation:
BBa_K2656011 is the RBS B0034 standardized into the Golden Braid assembly method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure correct spacing for the CDS expression when assembled into a TU.
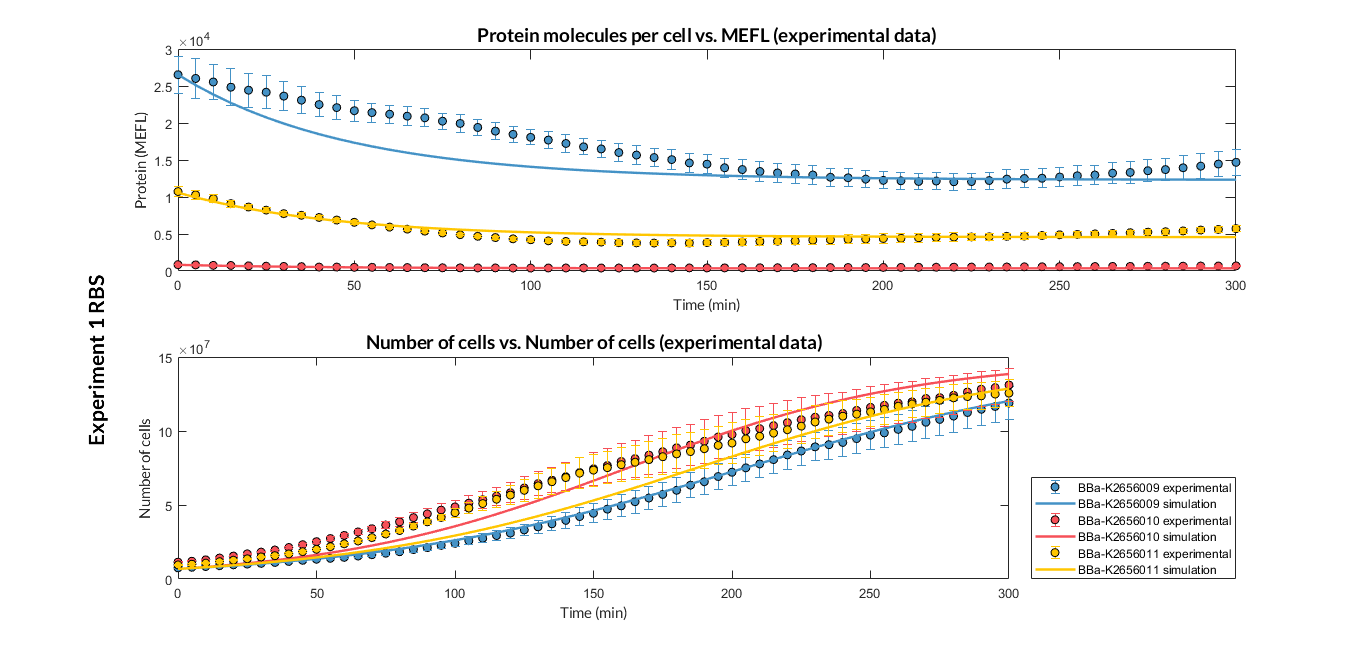
Characterization of the this part was performed with the transcriptional unit BBa_K2656101 , which was used in a comparative RBS expression experiment with composite parts BBa_K2656101 and BBa_K2656102. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its [http://2018.igem.org/Team:Valencia_UPV/Modeling#optimization optimization values] based on each RBS tested.
| Table 1. Optimized parameters from experimental data (BBa_K2656009 RBS). | |||
| Parameter | Value | ||
| Translation rate p | p = 0.082 min-1 | ||
| Dilution rate μ | μ = 0.01631 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656008 (GB BBa_B0032 RBS) relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.371 | ||
| p parameter ratio (pRBS/pref) | 0.398 | ||
Shanghai_HS_United 2019's characterization
BBa_B0034 RBS (Elowitz 1999) -- defines RBS efficiency
All the construction is type IIS standard assembly. The different scar may effect the RBS efficiency for expression.
culture
Material: LB medium (2mL), pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, pSB1K3-GFP, pSB1K3-LacZ (BBa_K3136014, BBa_K3136015, BBa_K3136017, BBa_K3136007)
1. Take several test tubes and add to 2mL Kan medium respectively.
2. Take the cultured pSB1K3-GFP, pSB1K3-LacZ, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, use the needle tip to take a small amount, and put it onto the shaking table.
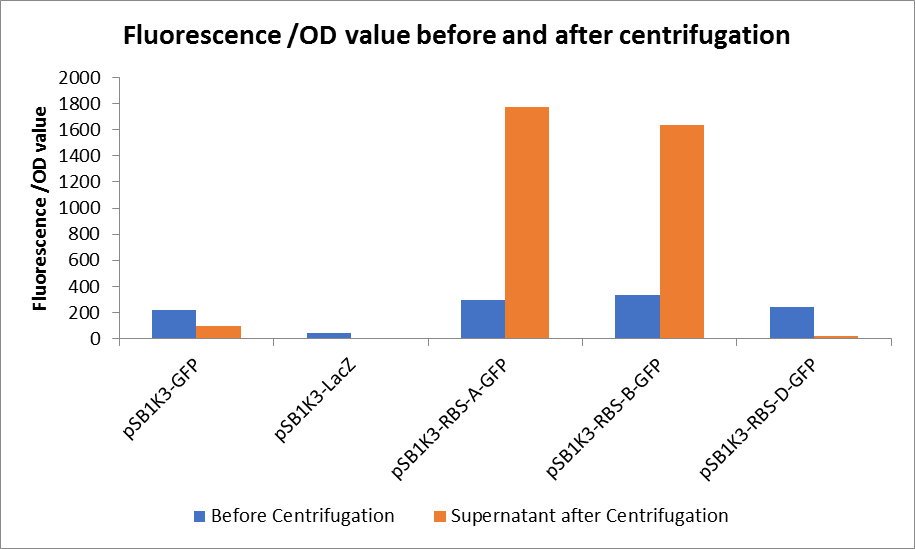
3. Measure the fluorescence value and the OD value before and after centrifugation.
Results:
In this experiment, RBS (Elowitz 1999) -- defines RBS efficiency is same as pSB1K3-GFP.
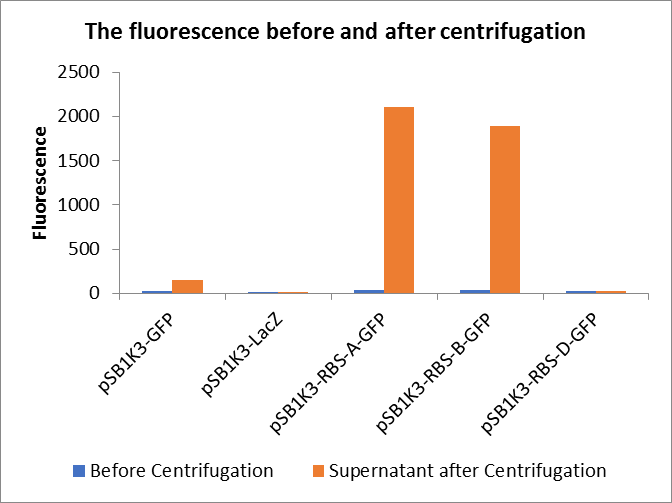
Through series of procedures, we have measured the fluorescence value before and after centrifugation (Fig1). The fluorescence of pSB1K3-GFP, pSB1K3-LacZ and pSB1K3-GFP, pSB1K3-LacZ have no differences whatever before or after centrifugation. But after centrifugation, the fluorescence of pSB1K3-RBS-A-GFP and pSB1K3-RBS-B-GFP all increased substantially.
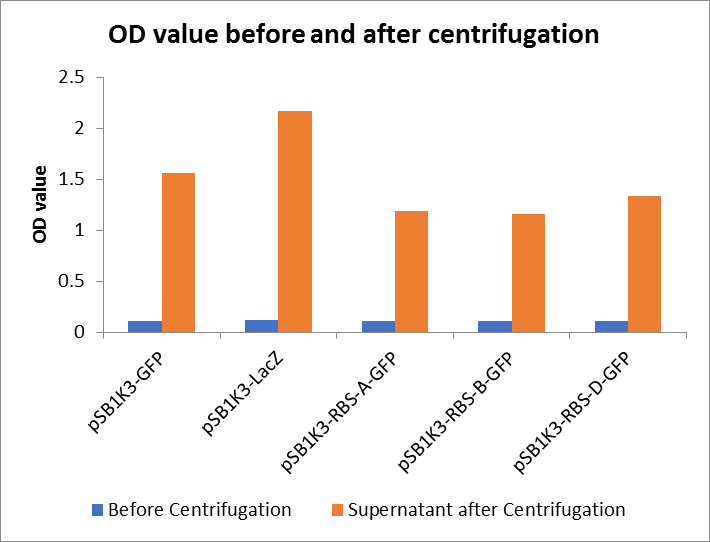
As shown in Fig2, the OD value of all samples before centrifugation was always lower than after centrifugation. Without the background interference of OD value, we can see the results clearly(Fig3). The fluorescence of pSB1K3-GFP hardly changed. It demonstrated that pSB1K3-GFP expressed a little. The intensity of RBS site in pSB1K3-GFP was weak.
(1) Protein Electrophoresis
Material: pSB1K3-LacZ, pSB1K3-GFP, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, loading solution (6 μL), deionized water (1000 μL), marker solution, Coomassie brilliant blue dye. Note: pSB1K3-LacZ, pSB1K3-GFP, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP are used for collaboration with the team Shanghai_HS_United, and these plasmids were expression in strain E. coli DH10B.
1. Add 24 μL pSB1K3-LacZ, pSB1K3-GFP, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, to 1.5mL tube respectively, and then add 6 μL loading.
2. Add 500 μL deionized water and centrifuge the tubes respectively.
3. Add another 500 μL deionized water, blow and suspend the solution.
4. Put the tubes 99℃ water for 15 minutes.
5. After heating, centrifuge the tubes for 5min.
6. Take and install the gel, add the electrophoresis buffer to the sample hole, and check if there is any leakage. Add marker, to the first and last sample holes, add pSB1K3-LacZ,pSB1K3-GFP and pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, MG1655 bpul, DH10B bpul, BL21(DE3) bpul to the other sample holes, respectively.
7. Connect the electrophoresis device to the power supply. Connect the positive electrode to the tank and the negative electrode to the slot for electrophoresis. The voltage is adjusted to 160V.
8. Turn off the power supply and disconnect the electrode until the bromophenol blue reaches the bottom of the release adhesive. Remove the glass sheet from the electrophoresis device and then remove the gel.
9. Soak the gel in Coomassie brilliant blue dye and dye it in a horizontal shaking bed for 15 min.
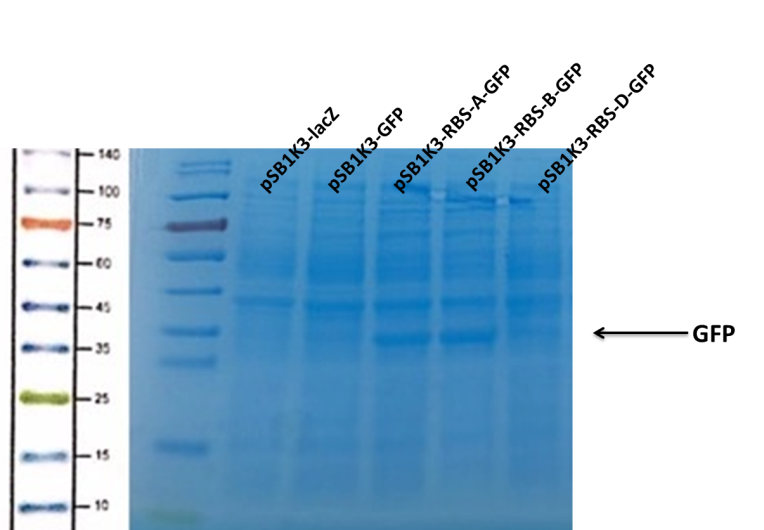
Observe the protein bands.
We can see that the lane of pSB1K3-GFP have no band(Fig4) at the location of GFP. It has been showed a low expression of pSB1K3-GFP and a feeble intensity of the RBS. Thus, the intensity of RBS was very important for the expression of proteins. The higher the intensity of RBS was, the more proteins expressed.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| biology | -NA- |
| efficiency | 1 |
//direction/forward
//function/coliroid
//rbs/prokaryote/constitutive/community
//regulation/constitutive
//ribosome/prokaryote/ecoli
| biology | |
| efficiency | 1 |

 1 Registry Star
1 Registry Star