Coding
ParoR
Part:BBa_K2984006
Designed by: Dimitri Schumacher Group: iGEM19_Humboldt_Berlin (2019-10-12)
Revision as of 21:41, 18 October 2019 by Chlamy Dima (Talk | contribs)
Codon-Usage-Optimized Paromomycin Resistance For Use in Chlamydomonas reinhardtii
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 298
Illegal NotI site found at 122 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 13
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 401
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Methods
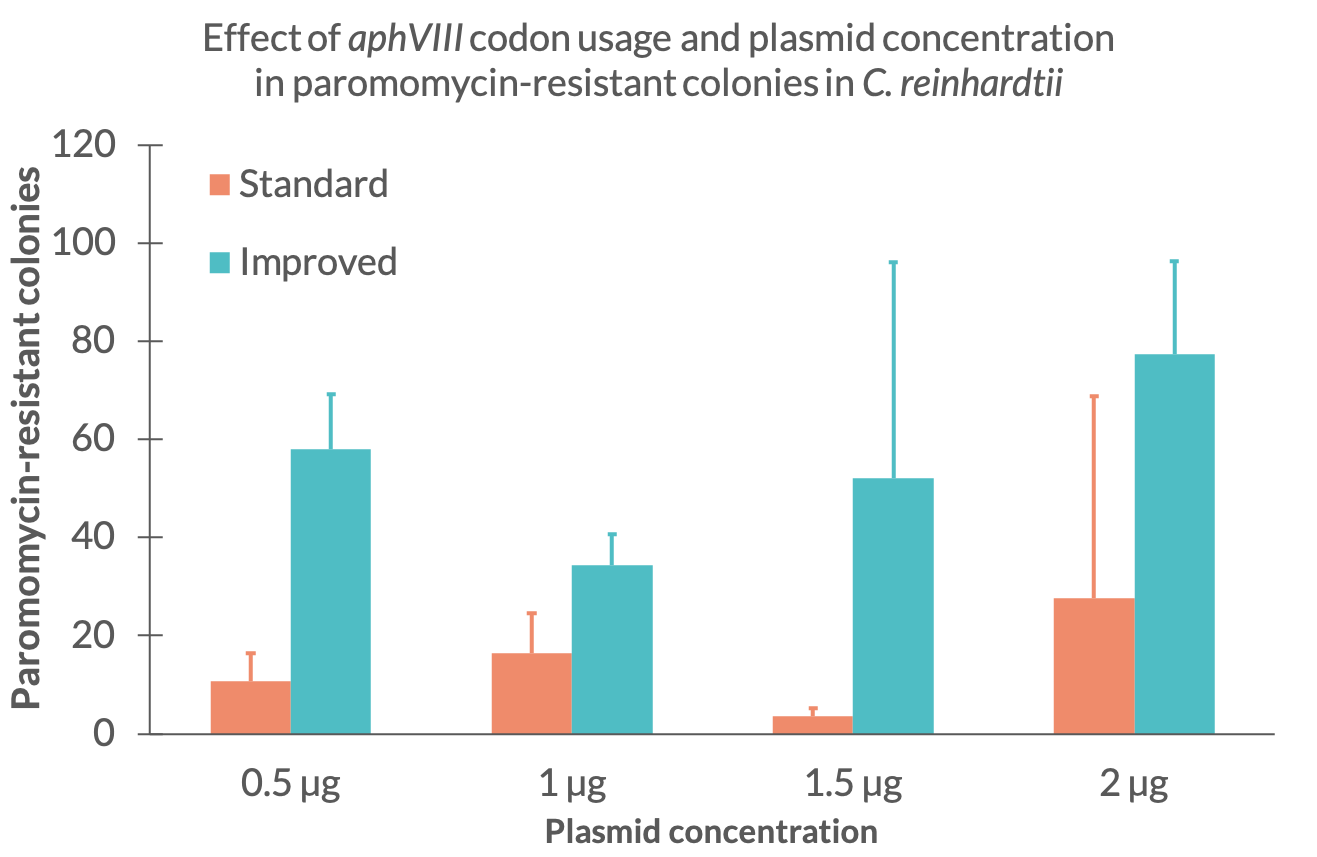
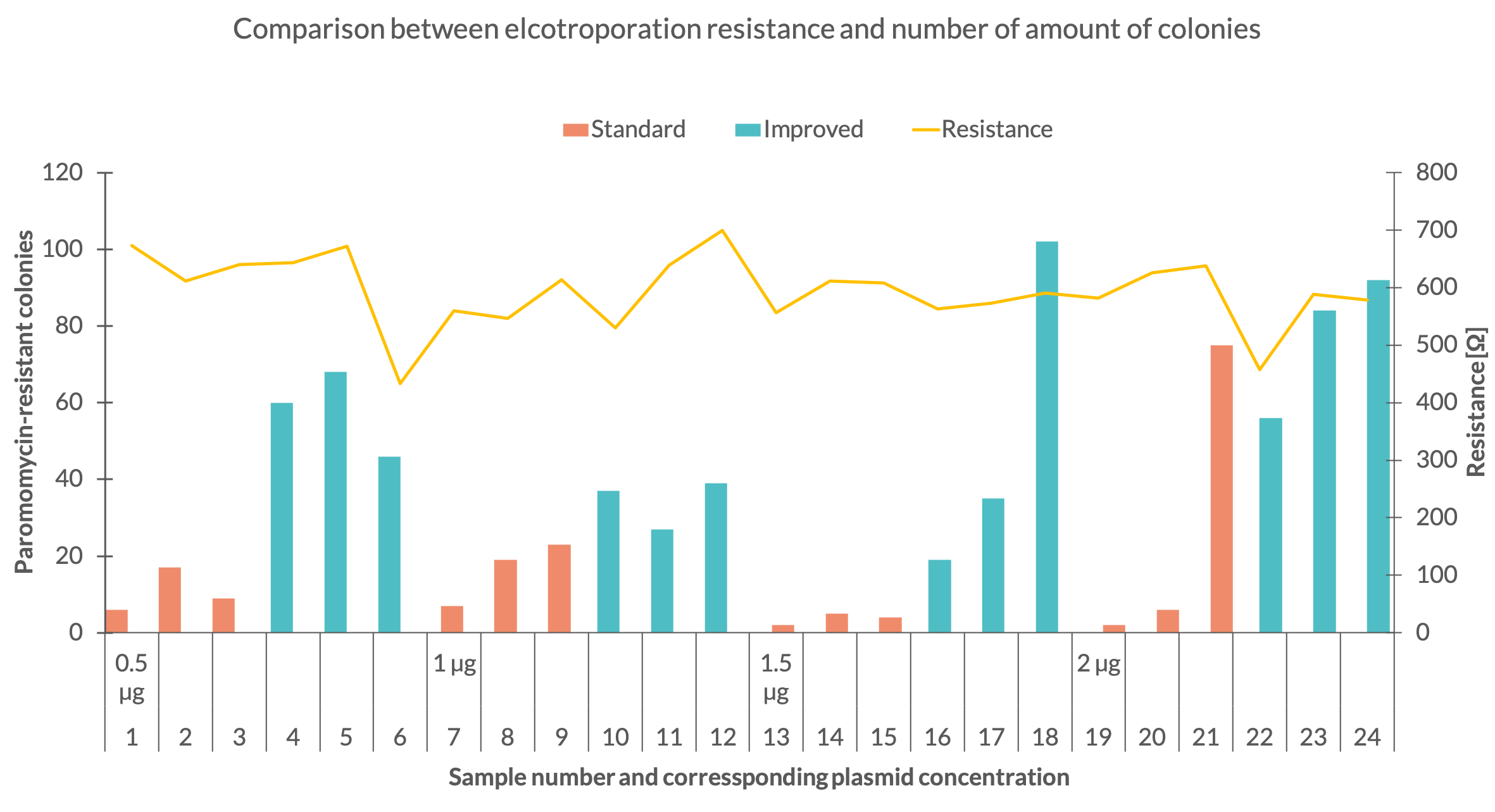
To see if the expression of the aminoglycoside 3’-phosphotransferase was increased, we performed several electroporations to transform C. reinhardtii with the improved paromomycin resistance. We used the C. reinhardtii strain UVM 4 since it is a strain designed to express transgene constructs (Neupert et al. 2009). We transformed the two paromomycin constructs with standard and improved codon usage starting with 0,5 µg DNA per electroporation sample and ascended with 0,5 µg steps up to 2 µg. For each construct and DNA mass we did three electroporations. The electroporation electrical resistance was measured for each sample. After resuspension and one day recovery in TAP medium, all samples were plated on TAP-agar plates containing a paromomycin concentration of 10 µM. After two weeks of growth, colonies corresponding to each sample were counted. Each colony of C.reinhardtii represents a successful transformation of the resistance and indicates the expression of the aminoglycoside 3’-phosphotransferase. By counting the amount of colonies on the plates, we could determine which construct and at which DNA mass at the time of transformation worked best.Results
First, we counted the total number of C. reinhardtii colonies that were transformed with the standard and improved resistance, regardless of the DNA mass used at the time of transformation. The results of the total colony count can be seen in Fig. 1. Counting the colonies we discovered that the total number of colonies was much higher for the improved plasmid version. The colonies of the samples using the standard usage resulted in a total amount of 175 whereas the improved plasmid version produced 665 colonies. This result indicates that the improved paromomycin resistance works better than the one with standard codon usage.
References
[1] Neupert, J., Karcher, D., & Bock, R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant Journal, 57(6), 1140-1150.
[2] Sizova, I., Fuhrmann, M., & Hegemann, P. (2001). A Streptomyces rimosusaphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene, 277(1-2), 221-229.
[edit]
Categories
Parameters
| None |
