Part:BBa_K1399007
GFP (mut3b) with SsrA-DAS+2 degradation tag
GFP (mut3b) (Part:BBa_E0040) with added engineered NYADAS-ssrA degradation tag (Part:BBa_M0051). The tag increases GFP turn-over rate, thus providing better temporal resolution of green fluorescence. In the same time, maximal fluorescence amplitudes will be lower as newly formed protein is degraded as soon as it is formed.
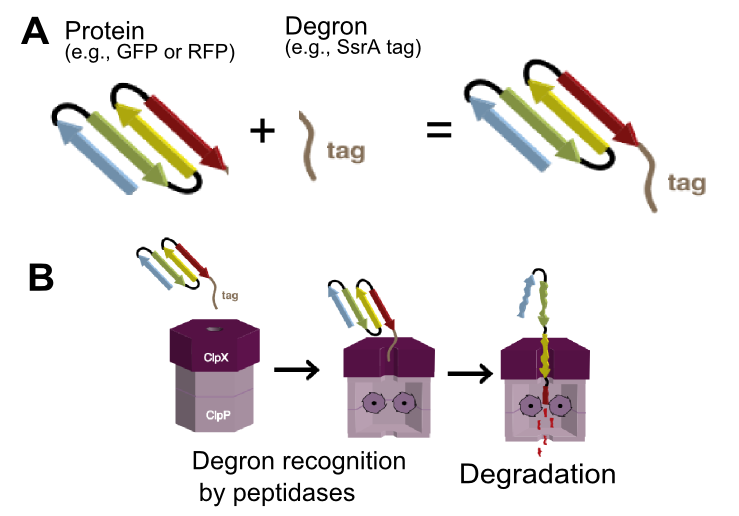
SsrA tags encode peptide sequence that is recognized by ClpA and ClpX unfoldases and ClpX mediator SspB (Figure 1).[1]
ClpA and ClpX then form a proteosome-like complex with ClpP protease and the protein is degraded.[1] The final three residues of the tag determines the strength of interaction with ClpX and thus the final protein degradation rate.[2] The NYADAS tag encodes peptide sequence AANDENYNYDAS is reported to have low affinity to ClpX thus its mediated degradation very much depends on the concentration of SspB (ClpX mediator).[1] The two additional residues ‘NY’ extends tag between SspB and ClpX binding sites, thus preventing clash when both proteins are bound to the tag.[3] However, be aware that exact protein degradation rate is influenced by multiple other factors: ClpXP and ClpAP protease concentrations, protein stability, Km of binding to the protease, temperature [4].
Contents
References
[1] Flynn, J. M. et al. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98, 10584–9 (2001).
[2] Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–6 (1998).
[3] McGinness, K. E., Baker, T. a & Sauer, R. T. Engineering controllable protein degradation. Mol. Cell 22, 701–7 (2006).
[4] Purcell, O., Grierson, C. S., Bernardo, M. Di & Savery, N. J. Temperature dependence of ssrA-tag mediated protein degradation. J. Biol. Eng. 6, 10 (2012).
2019 OUC-China's characterization
Background
This part was GFP(mut3b) followed by a variation of the WT SsrA tag sequence, which codes for the amino acid sequence AANDENYNYADAS. When the degradation tag fused to the C-terminal of proteins, it will make the protein susceptible to fast degradation through SspB-mediated binding to the ClpX protease as well as ClpA. DAS variants of SsrA tags require SspB for efficient degradation.
Note that tagged protein degradation rates are dependent on concentration of proteases (ClpX, ClpA) and binding mediators (SspB). Degradation rates are also heavily dependent on protein stability and depend on Km of binding to the protease, which is highly variable with each substrate. Be aware of ClpX, ClpA, and SspB knockouts when using SsrA tags; some expression strains have these mutations.
Result
Compared with GFP(mut3b), we found that the degradation tag has the ability to degrade the protein quickly.
Protocol
Extend to our project
This year, OUC-China has proposed a guideline to construct modular riboswitch for the future iGEM teams. The modular riboswitch we designed consists of the original riboswitch, Stabilizer and Tuner. Stabilizer can protect the structure of riboswitch from damage while Tuner have the ability to separate Stabilizer and GOI. The design principle can tackle many problems about riboswitch including context-dependent performance and limited dynamic range.
Furthermore, given that the accumulation of Stabilizer can lead to the increased metabolic pressure on cells, affecting cell function and the expression of target genes, we came up with a idea to help reduce intacellular space utilization. We constructed a Tunercontaining a degradation tag which can degrade Stabilizer. The results can demonstrate that this tuner can improve response curve of riboswitch successfully!
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 644
//chassis/prokaryote/ecoli
//function/reporter
//function/reporter/fluorescence
| color | Green |
| direction | Forward |
| emission | |
| emit | 511 |
| excitation | |
| excite | 501 |
| kegg | |
| lum | |
| protein | GFPmut3b |
| swisspro | |
| tag | SsrA-DAS+2 degradation tag |