Part:BBa_K2558203
Ptac test device with Anderson strong-expressed lacI
Tac promotor is a hybrid between trp and lacUV5 promoters. Tac promotor is a kind of strong E. coli promotors and is IPTG inducible. This device contains constitutively (BBa_J23100, Anderson strong promotor) expressed lacI protein and Ptac controlled sfGFP reporter. We designed this device along with K2558204 and K2558205, which are the same except for the strength of the constitutive lacI expression. We aim to specify the relation between the level of lacI production and the quality of IPTG induced expression.
Usage and Biology
In order to investigate how lacI dosage affect IPTG induction we used Anderson promotor J23100, J23110 and J23114 to design three constitutive lacI generator of different intensity. The three lacI generator were then ligated with Ptac controlled reporter sfGFP to make three IPTG induction devices (https://parts.igem.org/Part:BBa_K2558203, https://parts.igem.org/Part:BBa_K2558204, https://parts.igem.org/Part:BBa_K2558205). By measuring sfGFP fluorescence we tested how these devices react to IPTG.
Results
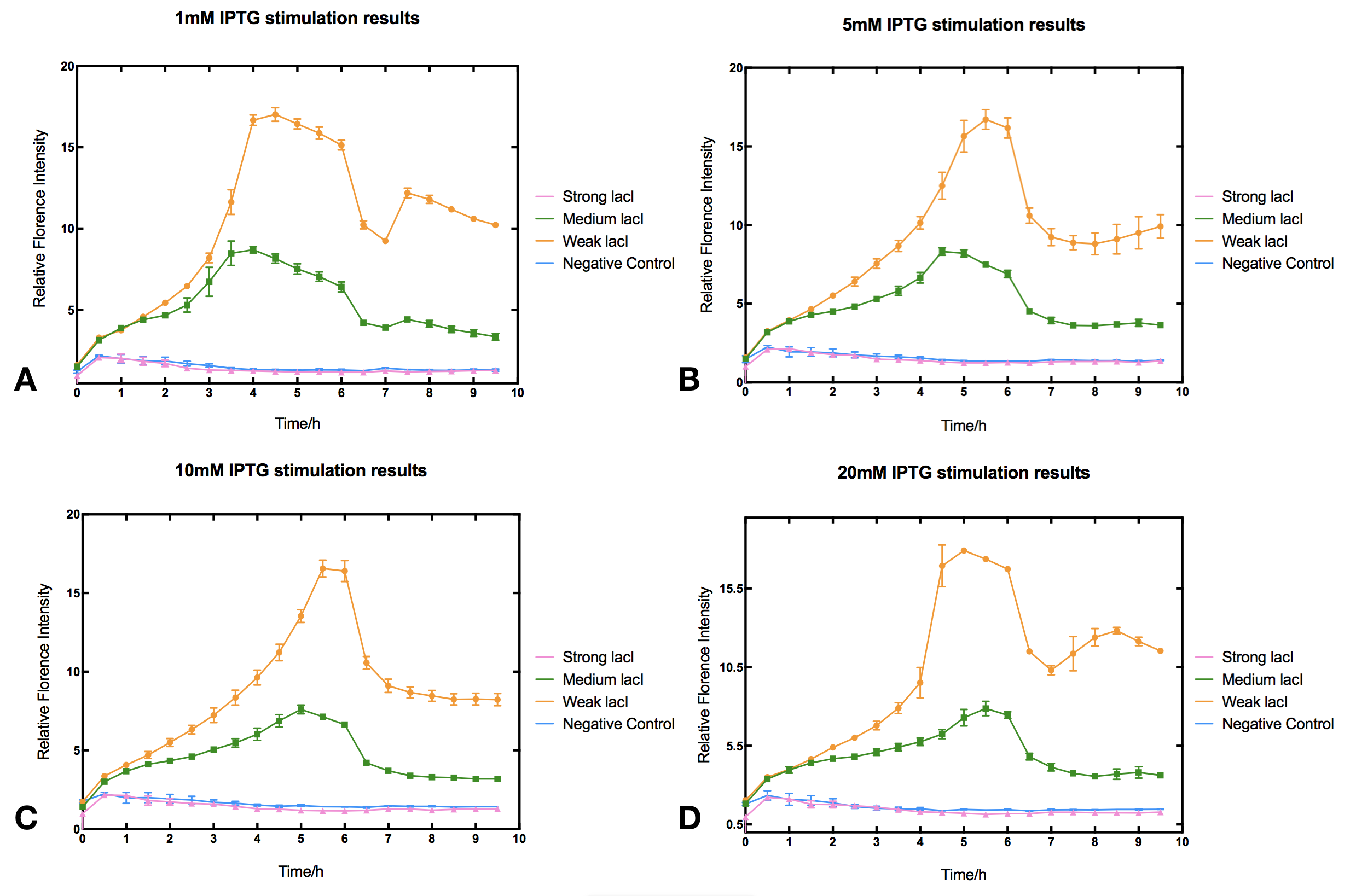
With high level of lacI expression (https://parts.igem.org/Part:BBa_K2558203), sfGFP fluorescence has almost no response to IPTG induction. Weak lacI expression (https://parts.igem.org/Part:BBa_K2558205) has the most significant IPTG induced sfGFP expression. With medium lacI concentration (https://parts.igem.org/Part:BBa_K2558204), the induction efficiency lies in between. Therefore, the result proves that high level of lacI expression severely decrease IPTG induction efficiency [1]. Furthermore, IPTG concentration can affect the regulation part performance. The figure shows that without IPTG the sfGFP florescence intensity will be at floor level. After IPTG addition, fluorescence signal immediately begins to climb, forming a peak at five hours after induction, then sfGFP florescence intensity will decrease and maintain at a lower level afterwards. IPTG concentration does not significantly affect the height of the peak or the expression level after the peak, but the peak width and expression stability of the system. Figures indicate that 5-10 mM IPTG has the most stable induction results.
Protocol
- one Transform the plasmids into E. coli BL21(DE3) or DH5α.
- two Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 5ml M9 medium with ampicillin at 100 ng/µl. Incubate for 6-8 h at 37℃ in a shaker.
- three Measure OD600 of the culture medium with photometer. Dilute the culture medium until OD600 reaches 0.6.
- four Add 100 µl bacteria culture medium into a sterile 96-well plate. Add IPTG to final concentrations of 0, 1, 5, 10, 20 mM. Fresh M9 medium serves as blank control. Positive control is colony constantly expressing sfGFP and negative control is colony without sfGFP expression. Place the 96-well plate into an automatic microplate reader. Incubate at 16℃ overnight and record the fluorometric value at 510 nm and OD600 for each well every 30 minutes.
- five Each group is repeated for at least 3 times.
Reference
[1] Szabolcs Semsey, Sandeep Krishna. "The effect of LacI autoregulation on the performance of the lactose utilization system in Escherichia coli" Nucleic Acids Res 2013 Jul; 41(13): 6381–6390
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1073
Illegal NheI site found at 1096 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2277
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 176
| None |
