Part:BBa_K2549041
NLS-TEVp
This part is one of the downstream elements of our amplifier. It is constructed by fusing NLS (Part:BBa_K2549054) and TEV protease (Part:BBa_K2549013), from N terminal to C terminal. NLS is a short nuclear location sequence from SV40 large T antigen. TEVp is a mutant TEV (tobacco etch virus) protease whose autoactivation is removed, thus performing more efficient cleavage[1]. When coexpressed with VP64-dNLS-ZF21.16 (Part:BBa_K2549039) in the same cell, it can destroy the nuclear localization sequence and prevent the transcription activator from inducing the gene expression. Similarly, the cleavage can be conducted with the KRAB-dNLS-ZF21.16 (Part:BBa_K2549040), thus keeping the repressor from switching off the gene expression.
Biology
TEV protease-based transcription regulation or proteolysis-only activity
TEV protease is widely used in synthetic biology for its high cleavage specificity (targetting amino acids sequence ENLYFQG/S between QG or QS)[2]. Rossner MJ et al have demonstrated the TEV activity-dependent activation of several reporters[3]. In our system, we set the TEV protease specific cleavage site into the spacer region of the nuclear localization sequence[4], which is the core design of the TEV protease-based complex logic gates.
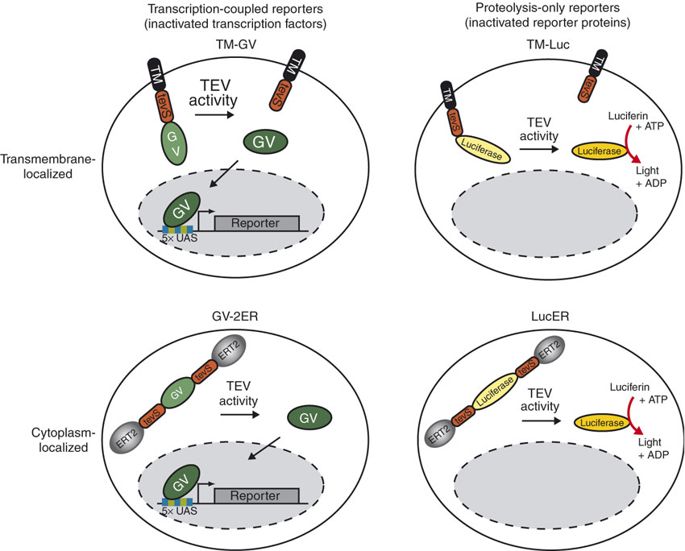
For more details about TEV protease, please refer to our TEVp (Part:BBa_K2549013).
Characterization
It works as we designed.
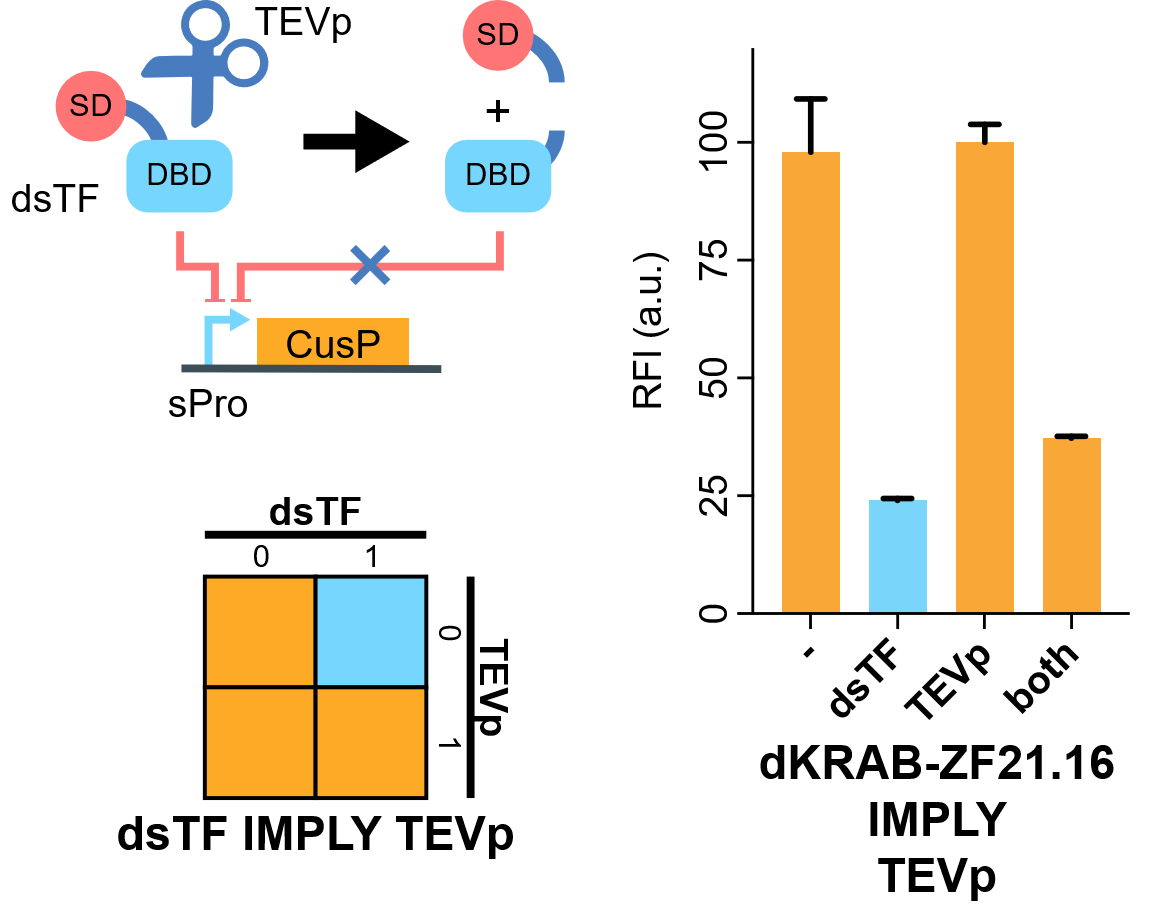
Flow cytometry results indicate that when KRAB-dNLS-ZF21.16 exists while TEVp doesn't exist that the expression level of d2EGFP is repressed. When TEVp and KRAB-dNLS-ZF21.16 coexist, TEVp cleaves dNLS and prevents KRAB from entering into the nucleus. Thus the repression is not severe as when KRAB-dNLS-ZF21.16 is present while TEVp is absent.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 337
References
- ↑ Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930
- ↑ Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Anal Biochem, 1994 Feb;216(2):413-7 PMID: 8179197; DOI: 10.1006/abio.1994.1060
- ↑ Monitoring regulated protein-protein interactions using split TEV. Wehr MC, Laage R, Bolz U, ..., Nave KA, Rossner MJ. Nat Methods, 2006 Dec;3(12):985-93 PMID: 17072307; DOI: 10.1038/nmeth967
- ↑ Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Robbins J, Dilworth SM, Laskey RA, Dingwall C. Cell, 1991 Feb;64(3):615-23 PMID: 1991323
//proteindomain/localization
| None |
