Part:BBa_K1639007
DAMP-Pexiganan
Synthetic 22-amino-acid antimicrobial peptide(AMP) pexiganan, which is a magainin AMP analog isolated from the skin of the African clawed frog.
Usage and Biology
Pexiganan, a 22-amino-acid antimicrobial peptide, is an analog of the antibiotic magainin peptides isolated from the skin of the African clawed frog has been shown to exhibit broad-spectrum microbicidal activity and acts with a bactericidal mechanism against which the likelihood of the development of resistance may be low. Furthermore Pexiganan molecules’ dose for destroying erythrocytes is 250 μg/ml, the other dose for destroying H.pylori is 16 μg/ml.(Figure 1)
A significant challenge in production of this antimicrobial peptide(AMP) in E.coli is it's intrinsic antimicrobial activity and proteolytic degradation of peptide during expression. To overcome this AMP is fused to carrier protein. DAMP4 is a protein formed by connecting, at the DNA level, four surface-active AM1 peptides. DAMP4 expresses at high levels of solubility in recombinant bacteria, and forms a four helix bundle structure that is thermostable to 90 C and remains soluble at relatively high salt conditions(Figure 2)

In our project we aim produce enough pexiganan molecules to eradicate all of the H.pylori population in the stomach. We inserted TEV protease cleavage site between DAMP4-Pexiganan to control releasing of Pexiganan. Due to the presence of H. pylori in the environment, extremely high concentrations of NH3 and Al2 molecules induce AND GATE the system (BBa_K1639000). As a result of this activation, TEV protease molecules will be produced. The produced TEV Protease break the large amount of connections accumulated between DAMP and Pexiganan in the cells and releases pexiganan molecules (Figure 5)
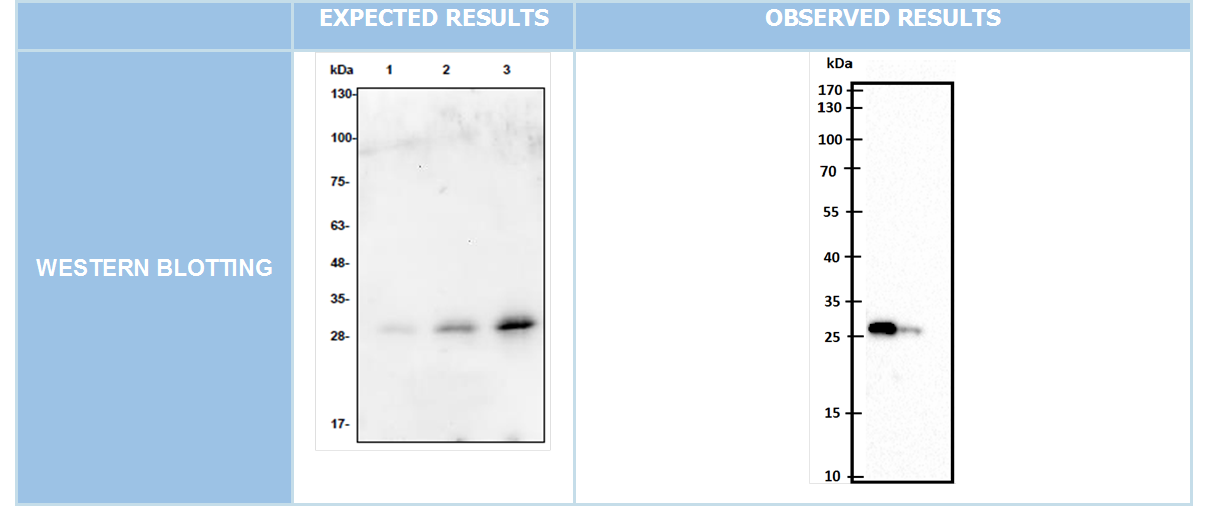
Cloning and Expression
We cloned our genes into pet45-b expression vectors(Figure 6) Expected bands must be at 699bp and 1041bp for DAMP4-PEX and TEV protease respectively. After succesfull cloning into expression vector Western blotting was conducted. (Figure 7-8)
To achieve cleavage of DAMP4-PEX complex by TEV protease both of them have to be expressed in the same bacterium but plasmids with same type of origin are not compatible. To overcome we cloned DAMP4-PEX into vector carrying ColA1 origin and TEV protease into pET45-b with ColE origin of replication.(Figure 9-10)
After cotransformation process we put bacterias at 37C for 16 hours incubation. End of 16 hours as a result we did not see any bacteria colony over agar plate. We retried several times this cotransformation process but with these tryings we havent seen any bacteria colony on our agar plates
//collections/antimicrobial
//collections/probiotics/production
| n/a | DAMP-Pexiganan |