Part:BBa_K1483002
α-Galactosidase
This part is [http://appft1.uspto.gov/netacgi/nph-Parser?Sect1=PTO1&Sect2=HITOFF&d=PG01&p=1&u=/netahtml/PTO/srchnum.html&r=1&f=G&l=50&s1=20140220553.PGNR. patented] and should therfore only be used for research purposes.
Encodes for an enzyme, that is capable of cleaving off galactose from B-group blood antigens, thereby turning them into H-antigens. Can be used to convert erythrocytes from blood type A to O. The part is designed in the RFC25 standard, thus enabling fusion to other proteins and tags.
Improvements from 2020 TAS_Taipei
We improved a part from the 2014 Tuebingen team: α-Galactosidase (α-Gal) from Elizabethkingia Meningoseptica (Part:BBa_K1483002).
Unsuccessful Expression of Past Part
We obtained the amino acid sequence of the α-Galactosidase protein from the iGEM DNA Repository Plate (BBa_K1483002), as entered into the iGEM parts collection database by the Tuebingen iGEM team in 2014.
In order to test protein expression of the enzyme, we added a strong promoter and strong ribosome binding site (RBS; BBa_K880005) upstream of the protein amino acid sequence to create a part BBa_K3717003, as well as a T7 promoter and strong ribosome binding site (RBS;BBa_K525998) upstream of the protein amino acid sequence to create a part BBa_K3717001.

Figure 2 - Colony PCR check for strong promoter (K88) α-Galactosidase (α-Gal) (Part: BBa_K3717003) using VF2 and VR primers. Uncut plasmid (K88 only control) has a band at the expected part size of 355 bp, indicated by white triangle. Confirms successful ligation as a band is produced at the expected size of 2107 bp, as indicated by the red triangle.
We tested protein expression of these two composite parts by transforming our plasmids into BL21 E. coli cells. We grew an overnight culture of the BL21 cells with our plasmids then diluted our cells to a standardized OD600 of ~0.1 and let it grow until an OD600 of 0.5~0.6. The diluted cultures of OD600 0.5~0.6 were then induced for expression with 0.5 M IPTG stock (to a final concentration of 0.5mM in the culture) and allowed to grow and induce overnight at room temperature.

Figure 3 - SDS Page of cell lysate for each strain: T7 promoter α-Galactosidase (α-Gal) (Part: BBa_K3717001) and strong promoter (K88) α-Galactosidase (α-Gal) (Part: BBa_K3717003). Green triangles indicate expected size for α-Gal (65.7 kDa). We found that the sequences for the target proteins do not contain a start codon, thus have no expression, as shown by the triangles.
Our SDS-page (Fig. 3) did not show any overexpression bands for the enzymes of interest. The results indicate that there were no target proteins at their expected band sizes: 65.7 kDa band for both T7 promoter + α-Gal and K88 promoter + α-Gal in the induced sample. As the SDS page is of cell lysis samples, other bands present are due to innate proteins present in the bacteria cell.
Upon comparison of the amino acid sequence from Tuebingen’s part (BBa_K1483002) with full sequences that were offered by other studies online, we discovered that the enzyme sequences were missing a few amino acids, also including the start codon, which explained the non-expression of the proteins (Fig. 4).

Figure 4 - Top sequence: First 24 amino acids of Team Tuebingen's 2014 α-Galactosidase part BBa_K1483002. Bottom sequence: First 60 amino acids of TAS_Taipei'sα-Galactosidase part BBa_K3717006; the first 15 amino acids include a 6x His tag and linker used for purification. Based on the alignment of the two sequences, Tuebingen's part is missing the first 21 amino acids of the α-Galactosidase protein.
Successful Expression of New Improved Part
We improved their part by adding the missing base sequences, including the start codon, and attached a 6x Histidine tag through a flexible Glycine-Serine linker. We derived the sequence of α-Galactosidase from Bacteroides fragilis then optimized the sequence for E. coli protein expression. This formed two new α-Galactosidase basic parts: α-Gal with N-Terminal 6x Histidine Tag (BBa_K3717006) and α-Gal with C-Terminal 6x Histidine Tag (BBa_K3717015).

Figure 5 - Open reading frame for α-Galactosidase with N-Terminal 6x Histidine tag (BBa_K3717006)

Figure 6 - Open reading frame for α-Galactosidase with C-Terminal 6x Histidine tag (BBa_K3717015)
We used these parts to create two new composite parts: BBa_K3717009 for N-Terminal His tag and BBa_K3717012 for C-Terminal His tag, through the attachment of a T7 promoter + RBS (BBa_K525998) upstream of the open reading frame (ORF) and a double terminator (BBa_B0015) downstream of the ORF.

Figure 7 - Design of α-Galactosidase with T7 Promoter, strong RBS, N-Terminal 6x Histidine tag and Double Terminator Construct (Part:BBa_K3717009)

Figure 8 - Design of α-Galactosidase with T7 Promoter, strong RBS, C-Terminal 6x Histidine tag and Double Terminator Construct (Part:BBa_K3717012)
We tested protein expression of the composite parts by transforming our plasmids into BL21 E. coli cells. We grew an overnight culture of the BL21 cells with our plasmids then diluted our cells to a standardized OD600 of ~0.1 and let it grow until an OD600 of 0.5~0.6. The diluted cultures of OD600 0.5~0.6 were then induced for expression with 0.5 M IPTG stock (to a final concentration of 0.5mM in the culture) and allowed to grow and induce overnight at room temperature.
We harvested the cells after the overnight induction and lysed them either through sonication or with xTractor Lysis Buffer (XTractorTM Buffer & XTractor Buffer Kit User Manual, n.d.) spiked with 500mM Imidazole stock (to a final concentration of 20mM in the lysate solution). We purified the Histidine tagged proteins using Ni sepharose affinity chromatography. We then utilized SDS-PAGE to confirm the sizes of purified proteins.
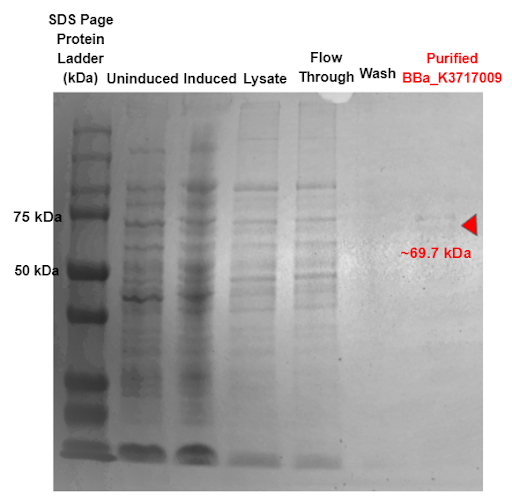
Figure 9 - SDS-PAGE of purified proteins with the T7 promoter α-Galactosidase expressing construct (BBa_K3717009). Red triangles indicate expected size for the part.
Our results in Figure 9 indicate a protein band at 69.7 kDa, which is the molecular weight of our α-Galactosidase ligase enzyme with the 6x His tag and GS linker attached. This shows that our α-Galactosidase (Part: BBa_K3717009) was expressed and purified.
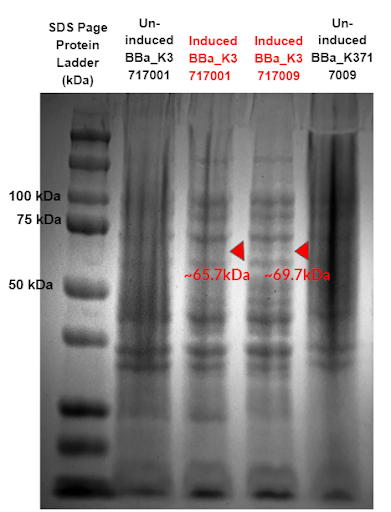
Figure 10 - SDS-Page comparing protein expression of two constructs using our α-Galactosidase sequence (BBa_K3717009) and Tuebingen’s α-Galactosidase sequence (BBa_K3717001). Red triangles indicate expected sizes for the parts.
Results from the gel (Fig. 10) show a band expressed at an expected size of 69.7 kDa for our new α-Galactosidase constructs, while no band expressed at an expected size of 65.7 kDa for Tuebingen’s α-Galactosidase constructs. This confirms our improvement of Tuebingen’s part BBa_K1483002 that lacked a start codon and thus was unable to express.
Successful Function of New Improved Part
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 412
| None |
