Part:BBa_K1379006
S. pneumoniae σx Generator
This is a protein generator for σX CDS (BBa_K1379004). The σX transcription is initiated by the strong constitutive promoter BBa_J23100 and the translation is initiated by strong RBS BBa_B0034, and transcription is terminated by BBa_B0015.
Usage and Biology
σX (ComX) is an alternative σ factor in Streptococcus pneumoniae which serve as a competence-specific global transcription modulator. (BioCyc) In S. pneumoniae, competence (a state capable of being genetic transformed) happens transiently during the log phase growth, and is regulated by a quorum sensing system utilizing the Competence Signal Peptide (CSP). (Luo & Morrison, 2003) Upon stimulation by CSP, σX will be expressed and associate with RNA polymerase apoenzyme. The resulting holoenzyme will then be guided by σX to initiate transcription of a set of “late” genes enabling genetic transformation and other unknown functions. Characterized genes regulated by σX were found to contain a 8-bp consensus sequence TACGAATA known as the Cin box or the Com box. (Piotrowski, Luo, & Morrison, 2009)
iGEM 2014 Hong_Kong_HKUST Team has cloned σX from S. pneumoniae strain NCTC7465 and characterized its ability to initiate transcription of two downstream promoters: PcelA(BBa_K1379000) and PcomFA (BBa_K1379001).
Following the principle depicted in Rhodius et al., (Rhodius et al., 2013) σX could achieve orthogonal gene expression regulation if used together with other sigma factors / ECFs. It should be noted that iGEM 2014 Hong_Kong_HKUST Team has compared the amino acid sequences of ECFs described by Rhodius et al. with that of σX from S. pneumoniae, and found no significant similarities, thus σX is a new sigma factor added to the list.
Characterization
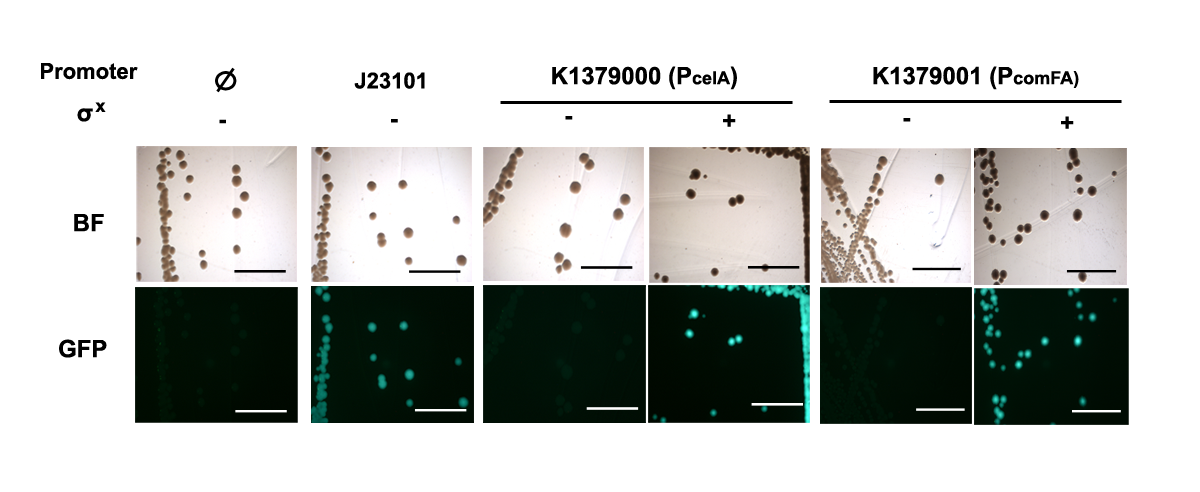
This generator, which expresses σX, was assembled with the promoter measurement kit BBa_E0240 with either promoter PcelA (Promoter only: BBa_K1379000, w/ BBa_E0240: BBa_K1379002) and PcomFA (Promoter only: BBa_K1379001, w/ BBa_E0240: BBa_K1379003). E. coli colonies holding the resulting constructs BBa_K1379005 and BBa_K1379007 in pSB3K3 were observed under fluorescent macroscope with UV filter. Measurement kit for standard reference promoter BBa_J23101, BBa_I20260 was used as a positive control; BBa_E0240 was used as the general negative control. for background fluorescence. Measurement kits for PcelA PcomFA without σX Generator were used as negative controls for function of σX.
As shown below, only in the presence of σX would PcelA and PcomFA be turned on, therefore, σX is functional.
Extensive documentation of our characterization can be found at our [http://2014.igem.org/Team:Hong_Kong_HKUST/pneumosensor/characterization characterization page for Pneumosensor].
Please refer to the pages BBa_K1379000 (PcelA) and BBa_K1379001 (PcomFA) for detailed characterization on the promoter activities in presence of σX.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 359
Reference
BioCyc was retrieved from http://www.biocyc.org/SPNE171101/NEW-IMAGE?type=GENE&object=GJC8-2079
Luo Ping, & Morrison Donald. (2003). Transient Association of an Alternative Sigma Factor, ComX, with RNA Polymerase during the Period of Competence for Genetic Transformation in Streptococcus pneumoniae. Journal of Bacteriology. doi:10.1128/JB.185.1.349-358.2003
Piotrowski Andrew, Luo Ping, & Morrison Donald. (2009). Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. Journal of Bacteriology. doi:10.1128/JB.01750-08
Rhodius Virgil., Segall-Shapiro Thomas., Sharon Brian., Ghodasara Amar., Orlova Ekaterina., Tabakh Hannah., . . . Voigt Christopher. (2013). Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Molecular Systhetic Biology. doi:10.1038/msb.2013.58
//chassis/prokaryote/ecoli
| None |
