Part:BBa_K1150024
uniCAS Histone Modifier (CMV promoter)
| CMV:dCas9-G9a | |
|---|---|
| Function | DNA binding protein fused to a methyl histone transferase |
| Use in | Mammalian cells |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Organism | Streptococcus pyogenes, Mus musculus |
| Source | Feng Zhang, Addgene Albert Jeltsch, University of Stuttgart |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
This device combines the dCas9 protein with the SET-domain of the murine histone methyltransferase G9a. dCas9 enables not only specific, but also multiple targeting of any DNA sequence(s) of interest. Hence, coupling of dCas9 to the effector G9a allows for specific methylation of histone H3 lysin 9 at any desired and accessible locus. These G9a induced histone modifications render open gene loci to a transcriptionally inactive state. [1]
To ensure localization to its site of action the dCas9, fused to the G9a SET-domain via a short linker, was flanked by two nuclear localization signals (NLS). Additionally, to enable detection of the expressed protein, it was tagged by a HA-epitope. Utilizing the strong CMV promoter in combination with the BGH terminator this device allows for constitutive, mammalian expression.

Figure 1.: Complete overview on CMV:dCas9-G9a with all features.
Usage and Biology
H3K9 methylation is a hallmark of repressed transcriptional states. [2] Upon close contact the murine G9a-set domain transfers methyl groups to H3K9 leading to transcriptional repression. [3]
The dCAS9 protein can simultaneously be targeted to several DNA loci as it interacts with small RNAs thereby forming a complex that will interact with complementary DNA strands. It origins from the bacterial adaptive immune system of Streptococcus pyogenes called CRISPR. Hijacking this system leads to a whole new approach for multiple gene targeting.
The team Freiburg 2013 combined these two elements to create a transcriptional repressor that can be targeted to any desired locus of interest.
This approach offers new possibilities for applied as well as for fundamental research, such as tissue engineering, epigenetics and cancer research.
Functional tests
Expression
To ensure protein expression of the fusion protein HEK-293T cells were seeded into 6-wells at a density of 150,000 cells per well. 24 hours later they were transfected with this device.
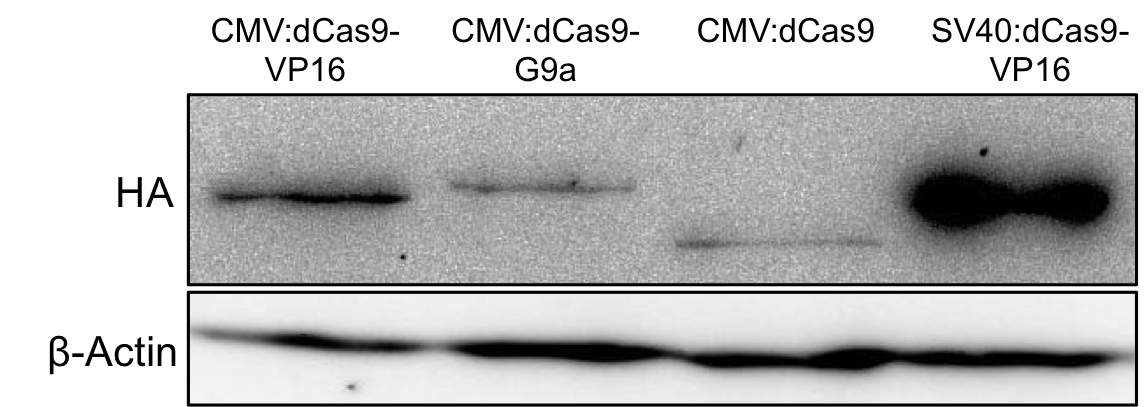
2 days post transfection the protein expression was evaluated by Western blot analysis of total cell lysates. Figure 2 demonstrates successful expression of the dCas9-G9a fusion protein by detection of the HA-tag.
Figure 2.: HA-tag fused to different dCas9-fusion proteins encoded on the RFC25 pSB1C3 backbone.
Proof of Function
Targeting of dCas9-G9a to an open, endogenous locus should lead to changes in the epigenetic state and repression. To test this, the endogenous VEGF-a locus was chosen, as it is well characterized and open in HEK-293T cells. Additionally, VEGF production can easily be measured by ELISA analysis.
Cells were seeded into 24-well format at a density of 65,000 cells per well and were co-transfected with the desired crRNAs, based on the RNAimer. These plasmids are derivates of the RNAimer, the only difference was the insertion of the target sites. These were: -8, -573, +434, -475 and -573/+434. As an off-target control crRNA targeting EMX was used. Every site was targeted with CMV:dCas9 wthout G9a, to differentiate the action of G9a from the CRISPRi effect. As an internal standard a constitutive SEAP reporter was used. 24 hours post transfection, VEGF levels and SEAP levels were measured and the ratio was calculated. Error bars represent standard deviations.
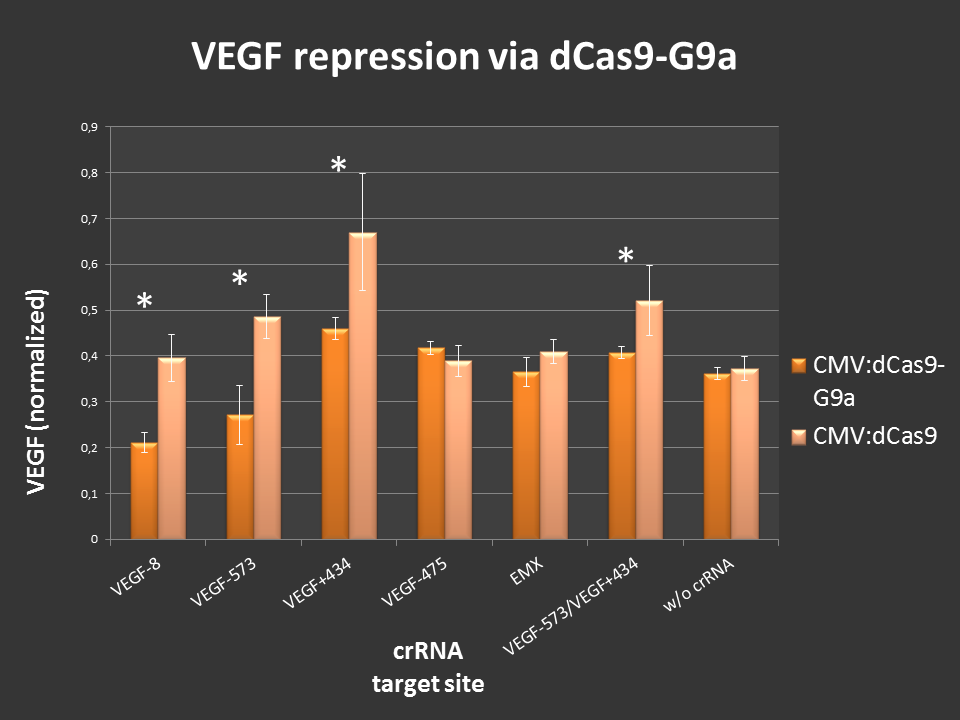
Figure 3.: dCas9-G9a targeting of endogenous VEGF-A locus in HEK-293T cells.
ELISA values, normalized to internal SEAP standard. For some loci clear repressive effects up to 50% were detectable. Controls did not display any off-target effects. Combining two target sites did not show any stronger repression.
Different VEGF target sites led to different repression efficiencies and simulatenous targeting of two sites did not further enhance the observed effects.
In summary, the CMV:dCas9-G9a is an effective repressor of endogenous gene expression, by modulating the chromatin structure of the targeted locus. This offers application in fundamental research as well as in medical science.
References
[1]Wolffe, A., et al. (1999). Epigenetics: Regulation Through Repression. Science 286169, 481.
[2]Snowden, A., et al. (2002). Gene-Specific Targeting of H3K9 Methylation Is Sufficient for Initiating Repression In Vivo. Current Biology 12, 2159-2166.
[3] Lee, D., et al. (2006). Histone 3 Lysine 9 Methyltransferase G9a Is a Transcriptional Coactivator for Nuclear Receptors. Journal of Biological Chemistry 281, 8476-8485.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 576
Illegal BglII site found at 900
Illegal BglII site found at 5375 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |