Part:BBa_K1159001
NanoLuc Luciferase in RFC[25]
NanoLuc Luciferase is engineered ATP-independent luciferase from a deep-sea shrimp which luminescense 2 magnitues higher than these from Renilla reniformis or from Photinus pyralis (firefly). Also the molecular weight of NanoLuc luciferase is twice smaller compared to other luciferase (only 19 kDa).
Usage and Biology
Experimental Data
Production in E. coli and purification
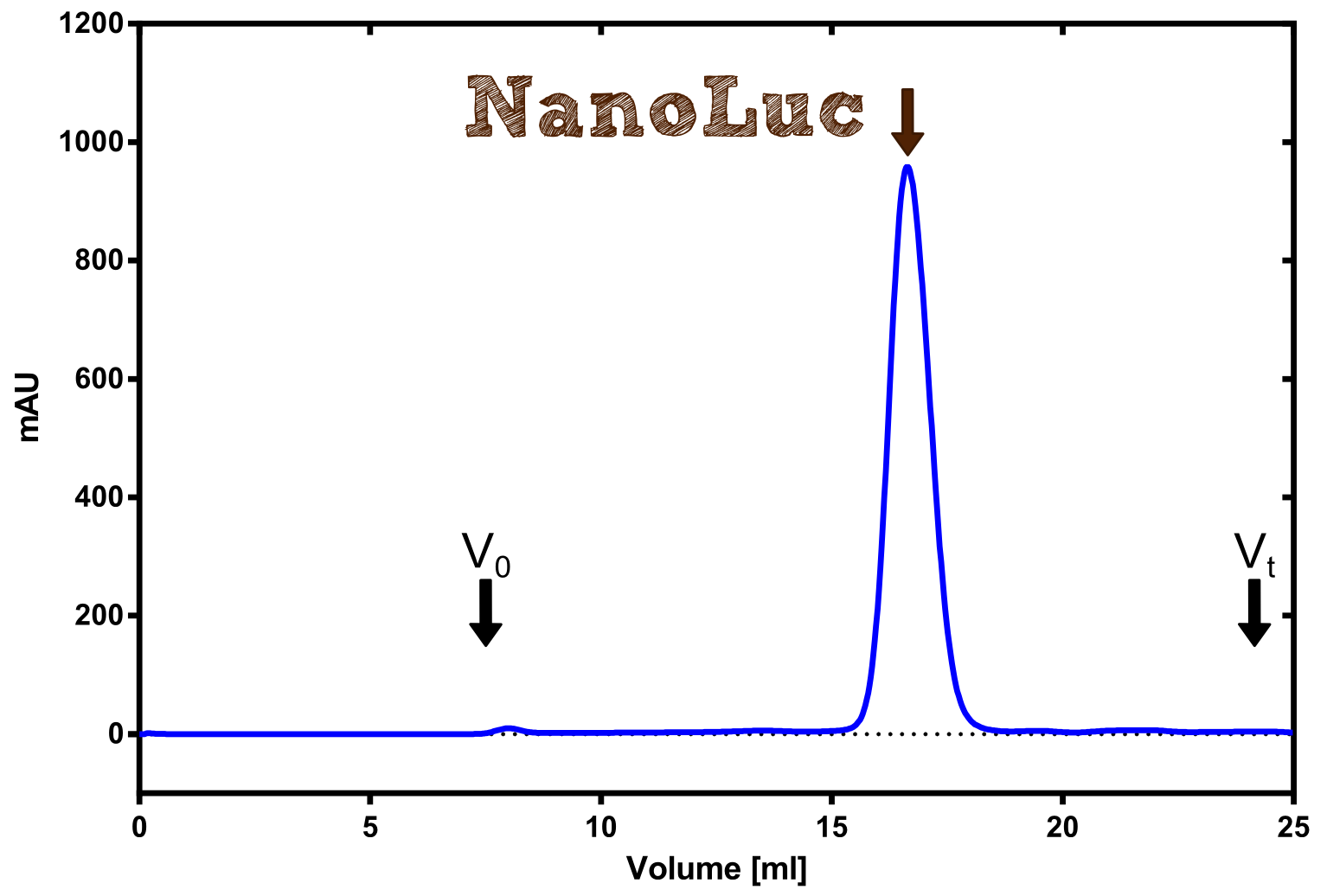
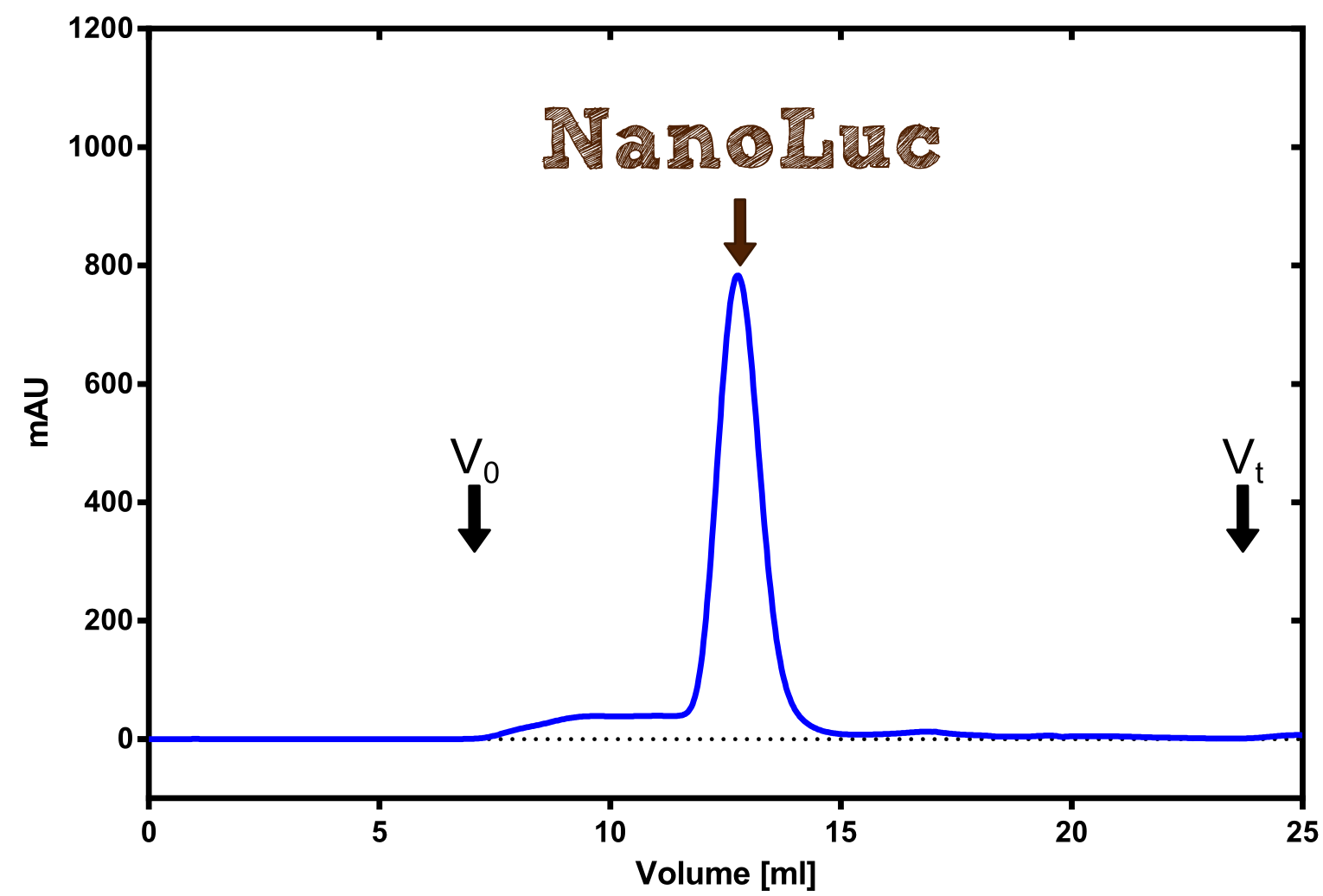
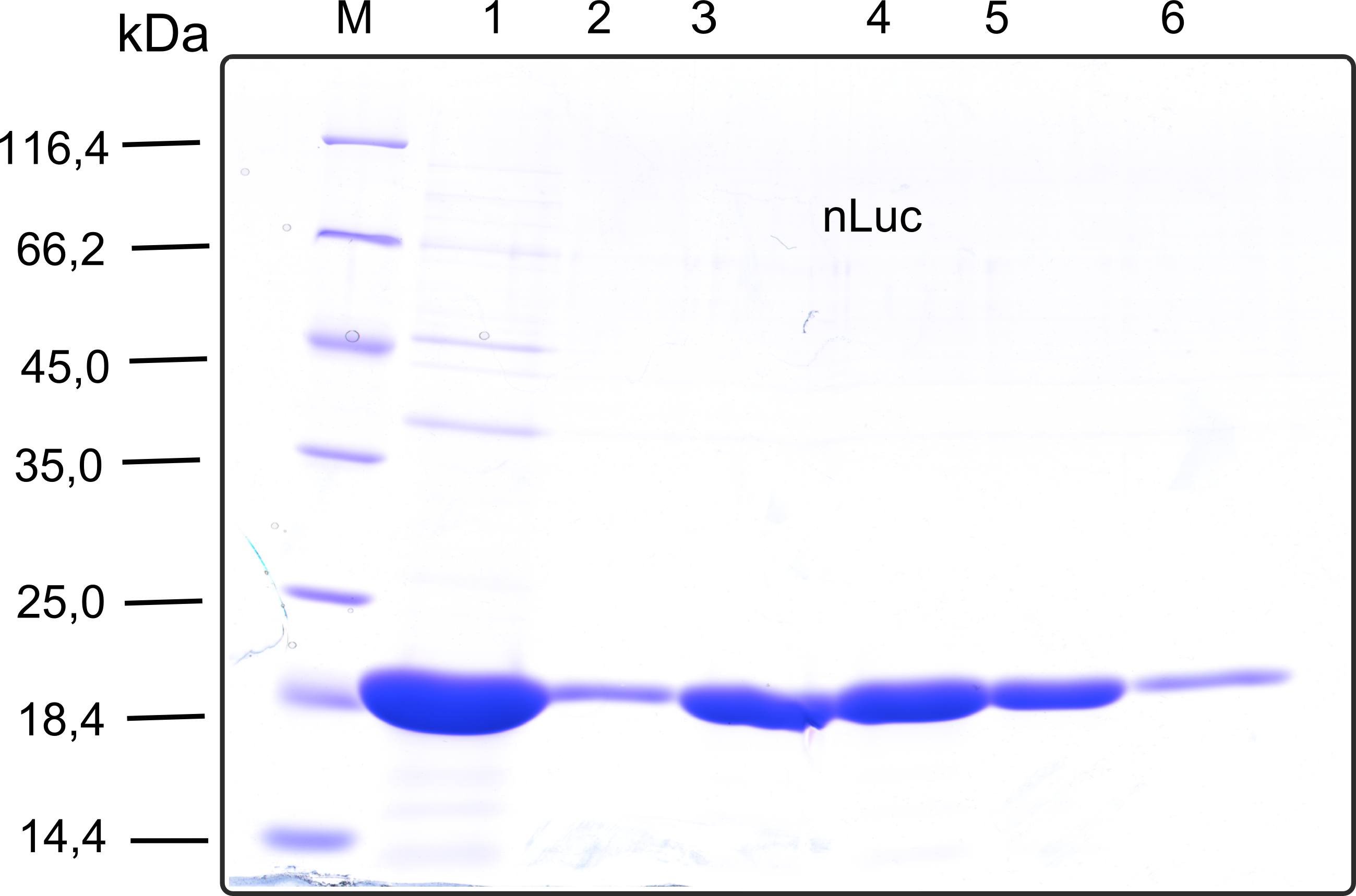
Therefore the NanoLuc was synthesized as a BioBrick in RFC[25] and was produced in E. coli using the pBad expression system with a C-terminal Strep-tag. After the production (2 l of LB-media for analytical and 12 l for preparative preparations) the cells were disrupted using sonification and the lysate was dialysed against 5 l of 1x SA-buffer. Afterwards the lysate was applied to a Streptavidin-Affinity (SA) column and was subsequently washed using SA-Buffer until a baseline was reached and the protein was then eluted using 5 mM of biotin (Attention: These are special columns which are not availible commercially. If you are using commercial colum material you have to use d-Desthiobiotin because usual biotin will elute your protein but you will not be able to regenerate the column after your chromatography). After the SA-chromatography the protein was concentrated using centrifugal concentration units (MWCO: 10 kDa). The concentrated protein was then applied on a Superdex S200/75 size exclusion chromatography. The chromatogram of both preparations show a single peak in the chromatogram which elutes at an expected elution volume of 15 ml. The absence of any notable aggregation peak shows the high stability of this protein and the ease of production.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 10
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
//chassis/eukaryote/ppatens
//function/reporter/light
| None |