Part:BBa_J04450:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Our aim was to evaluate RFP expression quantity of E. coli. In order to obtain this information, we agreed that the measurement of fluorescence intensity in certain OD values is the best approach. However, fluorescence quenching was a setback which we must overcome for accurate and reliable results. Because, as a result of quenching, fluorescence intensity counts do not give the actual amount of fluorescence expected from a certain OD value. Therefore we employed a curve fitting method suggested by Zhang et. al (2010) which is claimed to take the quenching problem into consideration

Where If is the fluorescence intensity; OD is the cell density; y0 is the offset; A1 is the amplitude; and t1 is the decay constant.
We chose BBa_J04450 for this experiment. And our negative control was again BBa_B0034.
There were nine bacterial concentrations prepared for one to five hours of IPTG induction. (All concentrations were prepared as triplicates.) Then a complete graph of all the induction durations was plotted with the help of the function shown above:

As seen in the graph, fluorescence behaves exactly as expected and does not increase linearly with the increased bacterial concentration. However, IPTG induction effect is clear on fluorescence intensity. (Error bars were too small to be visible.)

Finally, the variables A1 and t1 of the fittings of each induction duration were used to obtain a mean fluorescence intensity (MFI) value:



By conducting this experiment, we were able to characterize RFP expression pattern of the part BBa_J04450 in the host Top Ten. Theoretical mean fluorescence intensity reflects the fluorescence quantum efficiency of the fluorophore RFP, and was found to be unrelated to the cell concentration. Since it is obvious from the results we have obtained that change in the OD does not correlate linearly with the change in the fluorescence intensity. Therefore this model proposes a solution to the unpredictability of the fluorescence amount coming from a certain bacterial population.
The characterization was performed by METU-TURKEY 2010 IGEM TEAM.
Applications of BBa_J04450
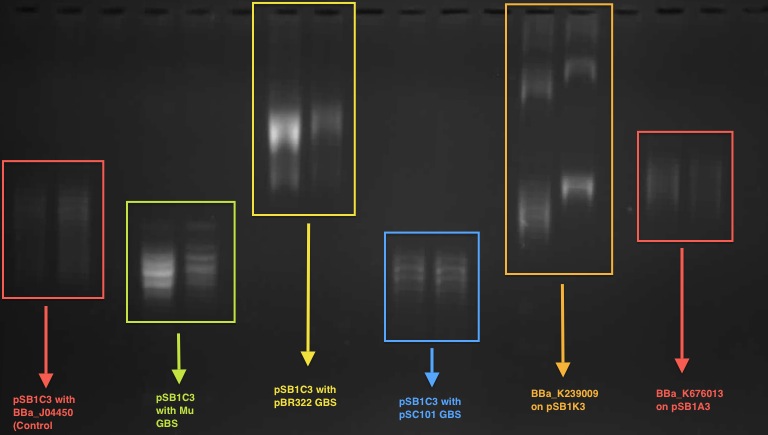
According to our result in the 1D chloroquine gel electrophoresis (at 2.5 μg/mL), the plasmid with the BBa_J04450 (on pSB1C3 backbone) migrated further compared to a plasmid with a known GBS (from pBR322 plasmid - BBa_K676012) on pSB1C£. At the chloroquine concentration which we used, the more negatively supercoiled plasmid will migrate further while the more relaxed molecules will travel slower through the gel.
So this indicates that the device BBa J04450 might have a potential gyrase binding site which facilitates the introduction of negative supercoils into plasmid compared to pBR322 GBS.
This characterisation was carried out by the UCL iGEM team 2011.
It was found that the Red fluorescence of this protein was not consistently apparent when expressed in biofilm. Numerous attempts to transform this construct into E. coli Nissle 1917 cells in a biofilm forming assay were unsuccessful, however, the RPF expressed readily into E. coli Nissle 1917 cells when grown on an agar plate.
The antibiotic resistance was still carried across when the culture was inoculated into the biofilm assay indicating that the plasmid was still present. The cultures were shaken at room temperature overnight.
It was thought that the lack of fluorescence was caused by the anoxic conditions generated by the biofilm assay, which may have resulted in poor folding of the chromophore. These results show that RFP is not the most efficient reporter molecule for biofilm, we would recommend something like iLOV, a new biobrick submitted by the Glasgow iGEM team this year [1].
Glasgow iGEM team 2011
User Reviews
UNIQ849ebb4f5aee5cf5-partinfo-00000006-QINU
|
•••••
Antiquity |
This review comes from the old result system and indicates that this part worked in some test. |
|
•••••
|
We've used this part as an alternative to the suicide part p1010 (ccdB) for selecting against cells that have been transformed with a [http://ginkgobioworks.com/support/BioBrick_Assembly_Manual.pdf destination plasmid]. It worked perfectly, and if i might say, it looks smashingly in LB medium ;). |
UNIQ849ebb4f5aee5cf5-partinfo-00000009-QINU

 1 Registry Star
1 Registry Star