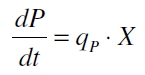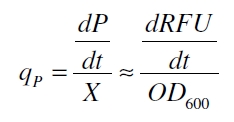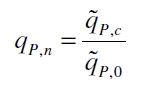Difference between revisions of "Part:BBa K389016:Experience"
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
| Line 5: | Line 4: | ||
===Applications of BBa_K389016=== | ===Applications of BBa_K389016=== | ||
| + | |||
| + | ====Data analysis for <partinfo>K389016</partinfo>==== | ||
| + | The data analysis is made in three steps. First step is the processing of the fluorescence raw data gained by the fluorescence plate reader for every sample: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_RFU_corrected.jpg|250px|center]] <div align="right">(1)</div> | ||
| + | |||
| + | |||
| + | In the second step the RFU<sub>corrected</sub> of every sample is plotted against the cultivation time it was drawn. The data is fitted by an exponential fit of the following style: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_Expfit.jpg|100px|center]] <div align="right">(2)</div> | ||
| + | |||
| + | |||
| + | The accumulation of mRFP in the cells is always exponential. A typical fitted product accumulation curve is shown below: | ||
| + | |||
| + | [[Image:Bielefeld_ExpFit_auf_RFU.jpg|500px|thumb|center|'''Exponential fit on the measured RFU plotted against cultivation time of a cultivation of <partinfo>K389016</partinfo> in ''Escherichia coli'' DB3.1 in LB medium with 10 mg ml<sup>-1</sup> chloramphenicol and 150 µM acetosyringone. ''']] | ||
| + | |||
| + | The product accumulation in a cultivation can be described as: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_Produktbildung.jpg|100px|center]] <div align="right">(3)</div> | ||
| + | |||
| + | |||
| + | with the amount of product P, the cell count X and the specific production rate q<sub>P</sub>. | ||
| + | |||
| + | RFU is commensurate to the concentration of mRFP (P) and the OD<sub>600</sub> is commensurate to the cell count (X) [1]: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_RFUpropP.jpg|100px|center]] <div align="right">(4)</div> | ||
| + | |||
| + | [[Image:Bielefeld_ODpropX.jpg|80px|center]] <div align="right">(5)</div> | ||
| + | |||
| + | |||
| + | With these assumptions it is possible to calculate the specific production rate of mRFP q<sub>P</sub> in the third step: the specific production rate for every sample of a cultivation is calculated by the derivation of the exponential fit line which describes the accumulation of product in the culture (dRFU/dt) and the measured OD<sub>600</sub> data: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_specific_production_rate.jpg|150px|center]] <div align="right">(6)</div> | ||
| + | |||
| + | |||
| + | The specific production rates q<sub>P</sub> of all samples of all cultivations made with a specific inductor concentration c are averaged and normalized against the specific production rate of the uninduced system q<sub>P,0</sub>: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_QPN.jpg|100px|center]] <div align="right">(7)</div> | ||
| + | |||
| + | |||
| + | This normalized specific production rate we calculated is commensurate to relative promotor units (RPU) which is commensurate to PoPS (polymerase per seconds) [1,2]: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_QPNRPUPoPS.jpg|150px|center]] <div align="right">(8)</div> | ||
| + | |||
| + | |||
| + | ====References==== | ||
| + | [1] https://parts.igem.org/Part:BBa_F2620:Experience/Endy/Data_analysis | ||
| + | [2] https://parts.igem.org/Part:BBa_J23101:Experience | ||
| + | |||
===User Reviews=== | ===User Reviews=== | ||
Revision as of 13:13, 23 October 2010
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K389016
Data analysis for BBa_K389016
The data analysis is made in three steps. First step is the processing of the fluorescence raw data gained by the fluorescence plate reader for every sample:
In the second step the RFUcorrected of every sample is plotted against the cultivation time it was drawn. The data is fitted by an exponential fit of the following style:
The accumulation of mRFP in the cells is always exponential. A typical fitted product accumulation curve is shown below:
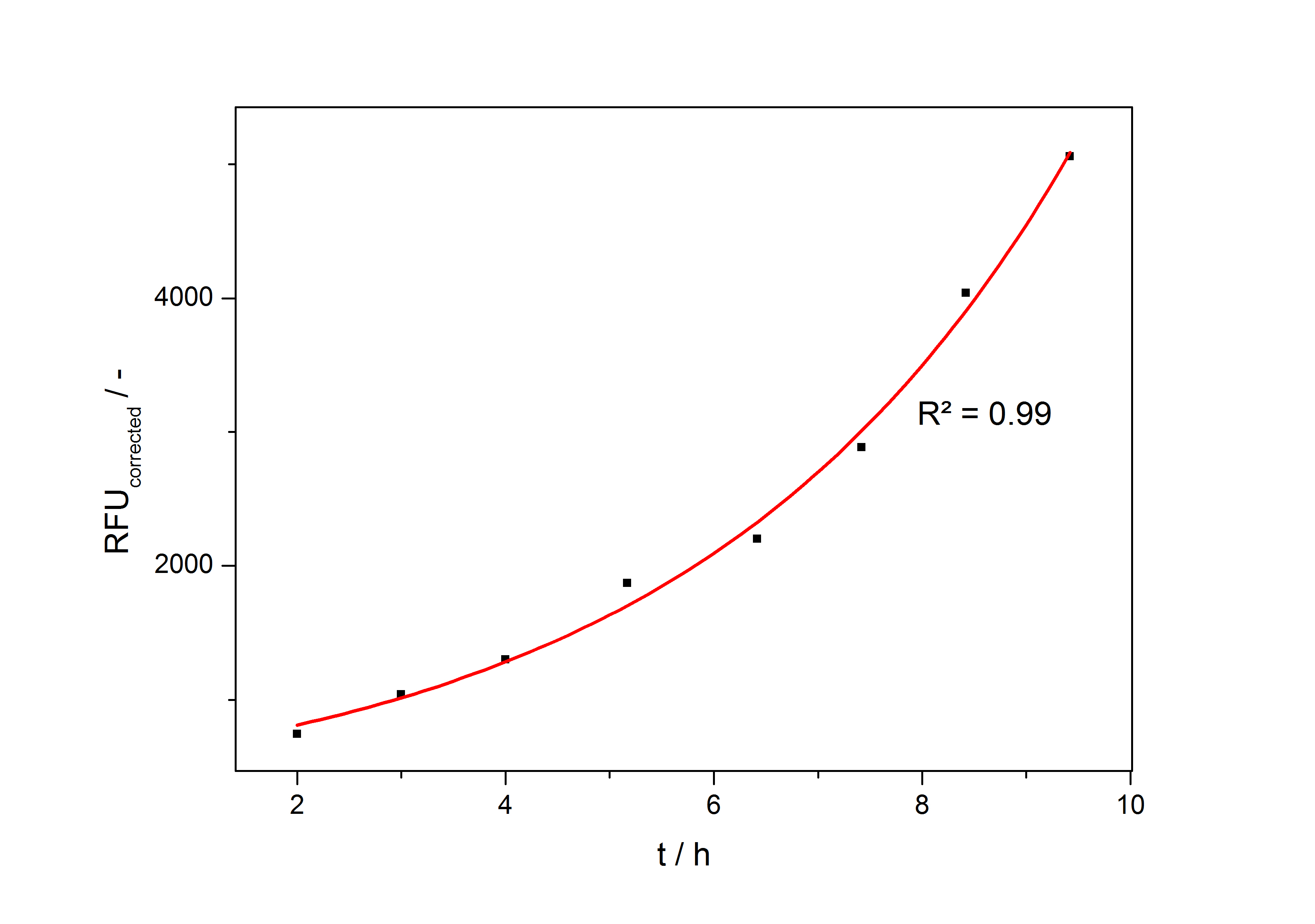
The product accumulation in a cultivation can be described as:
with the amount of product P, the cell count X and the specific production rate qP.
RFU is commensurate to the concentration of mRFP (P) and the OD600 is commensurate to the cell count (X) [1]:
With these assumptions it is possible to calculate the specific production rate of mRFP qP in the third step: the specific production rate for every sample of a cultivation is calculated by the derivation of the exponential fit line which describes the accumulation of product in the culture (dRFU/dt) and the measured OD600 data:
The specific production rates qP of all samples of all cultivations made with a specific inductor concentration c are averaged and normalized against the specific production rate of the uninduced system qP,0:
This normalized specific production rate we calculated is commensurate to relative promotor units (RPU) which is commensurate to PoPS (polymerase per seconds) [1,2]:
References
[1] https://parts.igem.org/Part:BBa_F2620:Experience/Endy/Data_analysis [2] https://parts.igem.org/Part:BBa_J23101:Experience
User Reviews
UNIQ4d1c3cd8dda44f2a-partinfo-00000002-QINU UNIQ4d1c3cd8dda44f2a-partinfo-00000003-QINU