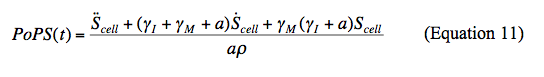Part:BBa_F2620:Experience/Endy/Data analysis
Data processing
Data processing consisted of two stages. First, we calculated the output of BBa_E0240 using the relative measurements of the plate reader. Second, we used this data and our knowledge of the behavior of BBa_E0240 to calculate the output of BBa_F2620
Raw data from the [http://openwetware.org/wiki/Endy:Victor3_plate_reader Wallac Victor3 multi-well fluorimeter] was processed by first subtracting the appropriate backgrounds. The absorbance of wells containing [http://openwetware.org/wiki/Endy:M9_media/supplemented supplemented M9 medium], Amedia, was subtracted from the sample absorbance data, Araw. The resulting data, Acorrected, was assumed to be directly proportional to the number of cells in the well.
Acorrected = Araw-Amedia ...(Equation 1)
Similarly, the fluorescence data for the GFP-free BBa_T9002 mutant, Gcells, was subtracted from the sample fluorescence data, Graw, and the resulting data Gcorrected was assumed proportional to the total number of GFP molecules in the well.
Gcorrected = Graw-Gcells ...(Equation 2)
The data was then converted to absolute units (cells/well and GFP molecules/well) using the calibration method described below. The conversion equations used are shown in Equations 3 & 4.
CFU = 3.1E8 Acorrected - 1.6E6 ...(Equation 3)
GFP = 7.0e8 Gcorrected + 6.0e11 ...(Equation 4)
Mean synthesis rates of GFP per cell, Scell, were calculated by assuming the total GFP synthesis rate, Stotal, to be equal to the time differential of GFP. Scell was then calculated via Equation 6. Note that since we have measured the total amount of GFP in the well and since we have assumed that GFP is not degraded, we can calculate the total synthesis rate of GFP and hence the per cell synthesis rate of GFP without considering dilution due to cell growth.
Stotal = d[GFP]/dt ...(Equation 5)
Scell = Stotal/CFU ...(Equation 6)
To interpret the behavior of BBa_F2620 from our observations of BBa_E0240 we employed an ODE model relating the output of BBa_E0240 to its input (the output of BBa_F2620. We defined the input to BBa_E0240 to be the time dependent rate of mRNA synthesis, PoPS(t) (mRNA per cell per sec). We defined the output of BBa_E0240 to be the synthesis rate of mature GFP, S_{cell} (GFP molecules per cell sec). The model includes two species and four parameters. The differential equations governing the levels of the two species are
d[M]/dt = PoPS(t) - γM[M] ...(Equation 7)
d[I]/dt = ρ[M] - (a+γI)[I] ...(Equation 8)
Scell = a[I] ...(Equation 9)
where [M] is the concentration of mRNA per cell, [I] is the level of immature GFP, PoPS(t) is the time dependent rate of mRNA synthesis, γM is the degradation rate of mRNA, ρ is the constant rate of protein synthesis per mRNA, a is the maturation rate of GFP, and γI is the degradation rate of immature GFP (incorporating degradation and dilution due to cell growth).
We parameterized the model using published and unpublished data and via experiments using BBa_T9002. We assumed a value of 4.8E-3 sec-1 for γM based on unpublished measurements performed in our lab on an almost identical mRNA, produced by BBa_E7107 (the transcript in the current study has two extra A nucleotides on the 5' end but an unchanged secondary structure). This value is also consistent with published data on mRNA decay in 'E. coli'. We assumed that dilution of mRNA due to cell growth was negligible relative to degradation.
We estimated a value for ρ of 0.4 proteins per sec per mRNA based on unpublished measurements from the Endy Lab on BBa_E7107. Again, this value is consistent with published data for translation rates per mRNA.
We measured an average GFP maturation rate of 1.8E-3 sec-1 as described above. Finally, we assumed that immature GFP is stable so that degradation was negligible relative to dilution due to growth. An average dilution rate of 2E-4 sec-1 was calculated from the multi-well fluorimeter absorbance data (corresponding to a 55 min doubling time).
At the 60 min timepoint used in the snapshot transfer functions, the above model for BBa_E0240 behavior is in steady state. Hence, we used the steady state relationship of Equation 10 to calculate the specific output of BBa_F2620 from the observed output of BBa_E0240.
PoPSss=γM(a+ γI)Ssscell/aρ ...(Equation 10)
To calculate the transient output of BBa_F2620 in the response time experiments, we rearrange the model (Equations 7, 8, and 9) to relate the time dependent PoPS output to measured values of Scell.
Data Calibration
Data measured using the [http://openwetware.org/wiki/Endy:Victor3_plate_reader Wallac Victor3 multi-well fluorimeter] was converted to absolute units to better allow repetition of the experiments and comparison of data from other researchers. Absorbance measurements were related to colony forming units (CFU) by growing a culture of MG1655 bearing BBa_T9002 to different cell densities and then plating an appropriate dilution on plates of [http://openwetware.org/index.php?title=LB&oldid=33777 Luria-Bertani medium] containing 20µg/ml kanamycin. For each culture density, three identical plates were made and the resulting colonies were counted. A straight line (R2 = 0.9912) was used to fit the data relating the number of colony forming units (CFU) per ml of culture to the absorbance measurements. The equation of the straight line was used to convert all absorbance measurements to CFU/well.
Fluorescence measurements were converted to an absolute number of GFP molecules by relating the fluorescence of cultures of MG1655 bearing BBa_T9002 to the fluorescence of purified GFP using a two-step calibration. In the first step, purified GFP (a generous gift from Chris Farrell) was serially diluted in cell lysate of wild-type MG1655. Lysis was performed using [http://www.piercenet.com/products/browse.cfm?fldID=3C6EACCB-232E-44DC-913F-6D9B16BEF8D5 B-PER II], a non-denaturing bacterial lysis solution (Pierce) using a modified [http://openwetware.org/index.php?title=Endy:Victor3_plate_reader/Calibrating_the_GFP-separated_label&oldid=52708 form] of the manufacturer’s protocol to ensure >90% lysis. The fluorescence of the GFP dilutions was measured in the [http://openwetware.org/wiki/Endy:Victor3_plate_reader Wallac Victor3 multi-well fluorimeter] and the data plotted against the mass of GFP in each dilution. A straight line (R2 = 0.99) was fit to the data. In the second step, cultures of MG1655 containing BBa_T9002 were induced using 1E-7 M 3OC6HSL for varying lengths of time leading to varying intracellular concentrations of GFP. Sample fluorescence was measured in the fluorimeter and the remainder of the cultures were lysed in an identical manner to that used for the purified GFP dilutions. The fluorescence of the lysed samples was measured and plotted against the fluorescence prior to lysis. A straight line (R2 = 0.97) was used to fit the data. The fit equations for the purified GFP dilutions and the lysed samples were combined to derive a straight-line relationship between the fluorescence of unlysed cultures and the number of GFP molecules/well. This relationship was used to convert all fluorescence measurements to GFP molecules per well.

 1 Registry Star
1 Registry Star