Difference between revisions of "Part:BBa K404201"
VolkerMorath (Talk | contribs) |
VolkerMorath (Talk | contribs) (→Cloning of Viral Bricks into capsid coding parts) |
||
| Line 58: | Line 58: | ||
All restriction endonucleases were purchased from [http://www.neb-online.de NEB].<br><br> | All restriction endonucleases were purchased from [http://www.neb-online.de NEB].<br><br> | ||
| − | <table border="1"> | + | <table border="1" style="float:right; width:480px; height:auto;> |
<tr> | <tr> | ||
<th><b>Restriction site</b></th> | <th><b>Restriction site</b></th> | ||
| Line 100: | Line 100: | ||
<html> | <html> | ||
| − | + | <div style="float:right; width:480px; height:auto; "><img src="https://static.igem.org/mediawiki/2010/5/55/Freiburg10_ViralBricks_function.png" width="460" | |
| + | height="auto"/></div> | ||
| + | <table border="1" style="float:right; width:480px; height:auto;> | ||
| + | <tr> | ||
| + | <th><b>Restriction site</b></th> | ||
| + | <th><b>Enzyme name</b></th> | ||
| + | <th><b>Recognition sequence</b></th> | ||
| + | <th><b>Activity in NEB-Buffer</b></th> | ||
| + | <th><b>Reaction temp.</b></th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>upstream 453</td> | ||
| + | <td>[http://www.neb.com/nebecomm/products/productR3132.asp SspI-HF]</td> | ||
| + | <td>[[image:Freiburg10_recognition site SspI-HF.Gif]]</td> | ||
| + | <td>25;100;0;100</td> | ||
| + | <td>37°C</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>downstream 453</td> | ||
| + | <td>[http://www.neb.com/nebecomm/products/productR3138.asp SalI-HF]</td> | ||
| + | <td>[[image:Freiburg10_recognition site SalI-HF.Gif]]</td> | ||
| + | <td>10;100;100;100</td> | ||
| + | <td>37°C</td> | ||
| + | <tr> | ||
| + | <td>upstream 587</td> | ||
| + | <td>[http://www.neb.com/nebecomm/products/productR3136.asp BamHI-HF]</td> | ||
| + | <td>[[image:Freiburg10_recognition site BamHI-HF.Gif]]</td> | ||
| + | <td>100;50;10;100</td> | ||
| + | <td>37°C</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>downstream 587</td> | ||
| + | <td>[http://www.neb.com/nebecomm/products/productR3151.asp PvuII-HF]</td> | ||
| + | <td>[[image:Freiburg10_recognition site PvuII-HF.Gif]]</td> | ||
| + | <td>0;25;0;100</td> | ||
| + | <td>37°C | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </div> | ||
<!---ViralBrick BAP---> | <!---ViralBrick BAP---> | ||
Revision as of 15:08, 17 October 2010
ViralBrick-453-BAP
| ViralBrick-453-BAP | |
|---|---|
| BioBrick Nr. | BBa_K404201 |
| RFC standard | RFC 25RFC 10 |
| Requirement | backbone without SspI, SalI, BamHI and PvuII |
| Source | synthetic |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
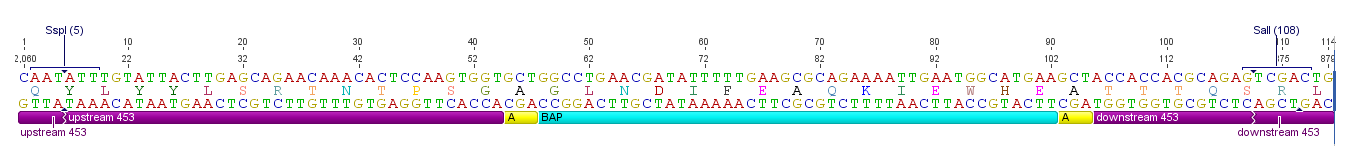
All capsid coding parts (e.g. (AAV2)-RepVP123) of the Virus Construction Kit designed by the iGEM Team Freiburg contain single cutting restriction sites on the side of the sequences coding for the two major surface exposed loops.
Using these restriction sites it is possible to insert functional motifs into the coding sequence for the viral capsid.
This BioBrick contains one of these functional motifs fanked by bilateral linkers and the viral sequences that code for the loop.
This category of BioBricks was termed ViralBrick to clarify that cloning does not function with the usual iGEM RFC restriction sites but with the mentioned ViralBrick restriction sites.
The four restriction sites for the ViralBrick insertion were designed in a way that the amino acid sequence of the viral capsid could be absolutely conserved. This was advisable because even slight changes in the viral capsid lead to drastically changed interactions with cellular surface receptors. For this reason we decided to insert these restriction sites in stead of using BioBrick assembly that would define at least two amino acids in the viral loop.
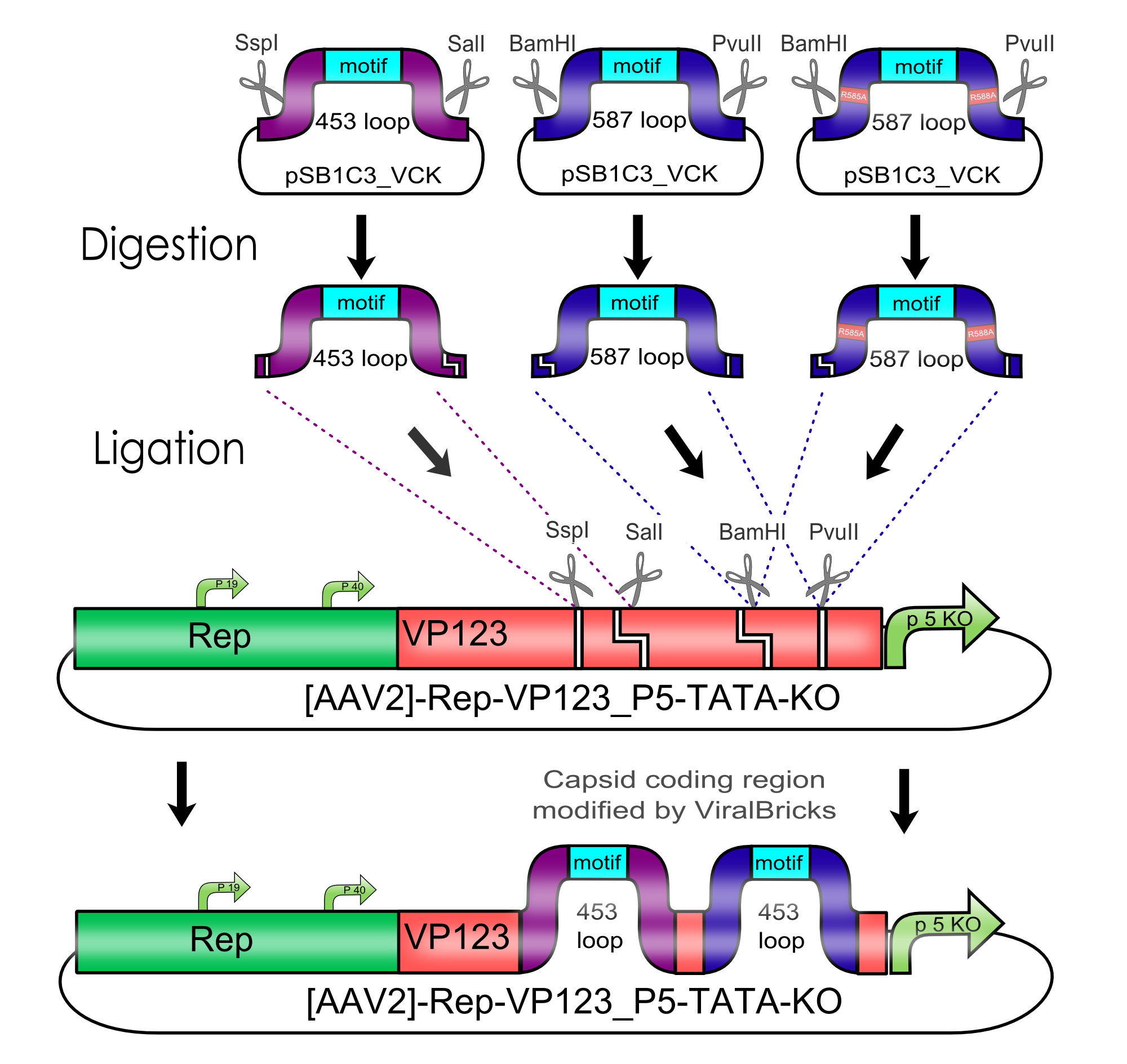
Modification of the Viral Capsid of the AAV2 using Viral Bricks
For therapeutical applications in human gene transfer the broad tropism for heparan sulfate proteoglycan (HSPG) has to be knocked-out and a novel tropism has to be inserted.
This retargeting can be realized either by insertion of functional motifs into the two major surface exposed loops or by fusion of these motifs to the N-terminus of the viral coat proteins.
The graphic on the right shows parts of the three-dimensional structure of a viral coat protein. The parts of the loop regions that are coded in the ViralBricks are shown in purple for the 453 loop and in blue for the 587 loop.
Cloning of Viral Bricks into capsid coding parts
In order to make loop insertions more convenient the following restriction sites were inserted into all capsid coding parts and already existing restriction sites were removed from the constructs.
The choice of these restriction sites was reasoned by enzyme performance, buffer compabilities and the number of existing restriction sites that had to be removed at other positions.
All restriction endonucleases were purchased from [http://www.neb-online.de NEB].