Difference between revisions of "Help:Spring 2010 DNA distribution"
(→Linearized Plasmid Backbone Instructions) |
|||
| Line 7: | Line 7: | ||
This year we have also included ready-to-use plasmid backbones for three of our standard plasmids: pSB1C3, pSB1T3, and pSB1A3. | This year we have also included ready-to-use plasmid backbones for three of our standard plasmids: pSB1C3, pSB1T3, and pSB1A3. | ||
| − | *'''<span style="color:red; font-weight:bold; font-size:115%;">NOTE:</span>''' The linearized plasmid backbones need to be cut by EcoRI and | + | *'''<span style="color:red; font-weight:bold; font-size:115%;">NOTE:</span>''' The linearized plasmid backbones need to be cut by EcoRI and PstI restriction enzymes prior to use. The label may incorrectly state that the backbone is pre-cut. |
==What's included with the Spring 2010 Distribution== | ==What's included with the Spring 2010 Distribution== | ||
| Line 30: | Line 30: | ||
* pSB1T3 | * pSB1T3 | ||
* pSB1A3 | * pSB1A3 | ||
| − | These plasmid backbones have been prepared via PCR and purified. All you need to do is cut with EcoRI | + | These plasmid backbones have been prepared via PCR and purified. All you need to do is cut with EcoRI, PstI and DpnI to leave two ends ready to be ligated to a Biobrick™ part. Use these backbones to send your parts to the Registry when shipping your parts before the Jamboree. |
| − | *'''<span style="color:red; font-weight:bold; font-size:115%;">NOTE:</span>''' The linearized plasmid backbones need to be cut by EcoRI and | + | *'''<span style="color:red; font-weight:bold; font-size:115%;">NOTE:</span>''' The linearized plasmid backbones need to be cut by EcoRI and PstI restriction enzymes prior to use. |
Revision as of 19:50, 29 June 2010
This year at iGEM we have made a few important changes to the Registry and the DNA distribution so please be sure to read through this information guide.
The Spring 2010 Distribution uses the same 384 well format used in 2007 and 2009, along with our extensive quality control measures (AB test plates, Growth plates, Restriction Digests, and Sequencing). While we will not be sending out all of the parts in the Registry, we have taken care to create a kit that includes both high-quality parts from the past as well as some of the 2009 submitted parts that were designated by teams as favorites. With a smaller number of confirmed quality controlled parts, (3) 384 well plates total, the Spring 2010 Distribution Kit will be a very effective basis for your synthetic biology projects.
This year we have also included ready-to-use plasmid backbones for three of our standard plasmids: pSB1C3, pSB1T3, and pSB1A3.
- NOTE: The linearized plasmid backbones need to be cut by EcoRI and PstI restriction enzymes prior to use. The label may incorrectly state that the backbone is pre-cut.
What's included with the Spring 2010 Distribution
- Spring 2010 DNA Distribution (3) 384 well plates
- Linearized Plasmid Backbones for pSB1C3, pSB1T3, and pSB1A3
- iGEM Stickers and Pins!
Getting Started with the Spring 2010 DNA Distribution
Storage
Since the distribution kit plates are comprised of dried DNA, they are quite stable for storage at room temperature. However once the DNA is resuspended in any of the wells, we recommend either storing the kit plate with its plastic cover in a -20C freezer, or aspirating the rest of the resuspended DNA from the well and keeping it separately in a -20C freezer.
The linearized plasmid backbones (25ng/ul at 50ul) should be stored at 4C or lower.
Usage
The Distribution Kit Plates contain dried DNA of hundreds of parts that are available in the Registry up to 2009. While there is not enough DNA for assembly, you will be able to transform the DNA into cells and then make your own glycerol stocks of any part that you wish.
Linearized Plasmid Backbone Instructions
In addition to the DNA plates the Distribution Kit also contains a set of linearized plasmid backbones:
- pSB1C3
- pSB1T3
- pSB1A3
These plasmid backbones have been prepared via PCR and purified. All you need to do is cut with EcoRI, PstI and DpnI to leave two ends ready to be ligated to a Biobrick™ part. Use these backbones to send your parts to the Registry when shipping your parts before the Jamboree.
- NOTE: The linearized plasmid backbones need to be cut by EcoRI and PstI restriction enzymes prior to use.
You can find instructions on how to use AND make your own linearized plasmid backbones on the protocol page.
DNA Kit Plate Instructions
To use the DNA in the Distribution Kit you may follow these instructions:
- With a pipette tip, punch a hole through the foil cover into the corresponding well to the Biobrick™-standard part that you want. Make sure you have properly oriented the plate. We recommend that you do not remove the foil cover, as it could lead to cross contamination between the wells.
- Add 10uL of diH2O (deionized water)
- Pipette 1 or 2uL of the resuspended DNA transform into your desired competent cells, plate bacteria with the appropriate antibiotic* and grow overnight.
- Pick a single colony and inoculate broth (again, with the correct antibiotic) and grow for 18 hours.
- Use the resulting culture to [http://openwetware.org/wiki/Miniprep/Kit-free_high-throughput_protocol miniprep] the DNA AND make your own glycerol stock (for further instruction on making a glycerol see [http://openwetware.org/wiki/Endy:Making_a_long_term_stock_of_bacteria this page]). We recommend using the miniprepped DNA to run QC tests, such as restriction digests and sequencing.
* To know which antibiotics to use, look at the plasmid that the part is in. The naming scheme for plasmids is specifically designed to indicate antibiotic resistance.
Note that there is not enough DNA to perform anything but transformations from the DNA in the wells.
DNA Kit Plate Orientation
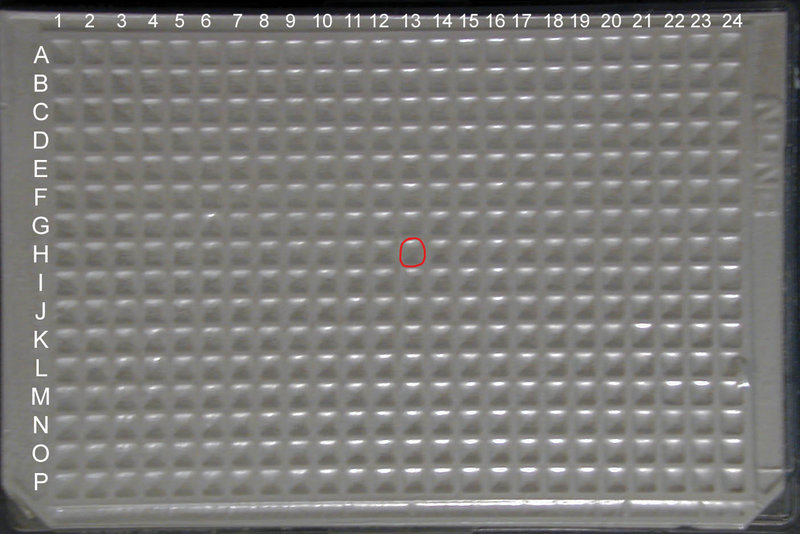
For the Spring 2010 Distribution, you will receive a set of (3) 384 well plates, from which you can extract the part of your choice once you have located it in the distribution. Unfortunately, the foil cover will obscure both column and well markings. However, you can still find your part by correctly orienting the plate using the two notched corners as markers: well A1 is located at the upper left corner of the plate when the long side of the plate with the notched corners is considered the bottom.
Once you have found the well which your BioBrick™ part of choice is located in by searching for it through the Registry (for example well 13H in iGEM 2009 DNA Parts Kit Plate 1) you want to count across the plate starting with Column 1 until you get to Column 13 and down the plate starting with Row A until you get to Row H.
MAKE SURE that the two notched corners of the plate are oriented at the BOTTOM of the plate (see the top view image at left for correct orientation)
Finding and Choosing Parts in the Spring 2010 Distribution
On the left-hand side of the Registry Main Page you will find a Catalog of Parts and Devices. This tool is meant to assist in the synthetic process through several avenues.
- It allows users to locate parts based on their function.
- It gives users the freedom to browse through different devices by type, through a series of tables which link to information in the parts registry.
- Parts and devices can be browsed by chassis. Unless otherwise specified, most parts in the Registry work in Escherichia coli.
- Parts and devices can also be browsed by standard. Unless otherwise specified, most parts in the Registry comply with the original BioBrick assembly standard (also known as Assembly standard 10).
- Parts and devices can be browsed by contributor.
- The chassis can be browsed to locate a particular strain.
Locating a Part in the Distribution
Prior to taking the DNA Distribution Kit out for use, it is helpful to know both the location of the part(s) you want, as well as the plasmid and antibiotic resistance(s) of the required parts. The Registry provides the necessary tools to locate your desired parts within the 384 well plates used in the distribution, along with antibiotic resistance and other pertinent information. If you would like to find a part to serve a particular function but have not yet picked it out, you can locate various parts by browsing, as described below. If, on the other hand, you already have a specific part number in mind, you can find its location by Focused Search.
Browsing
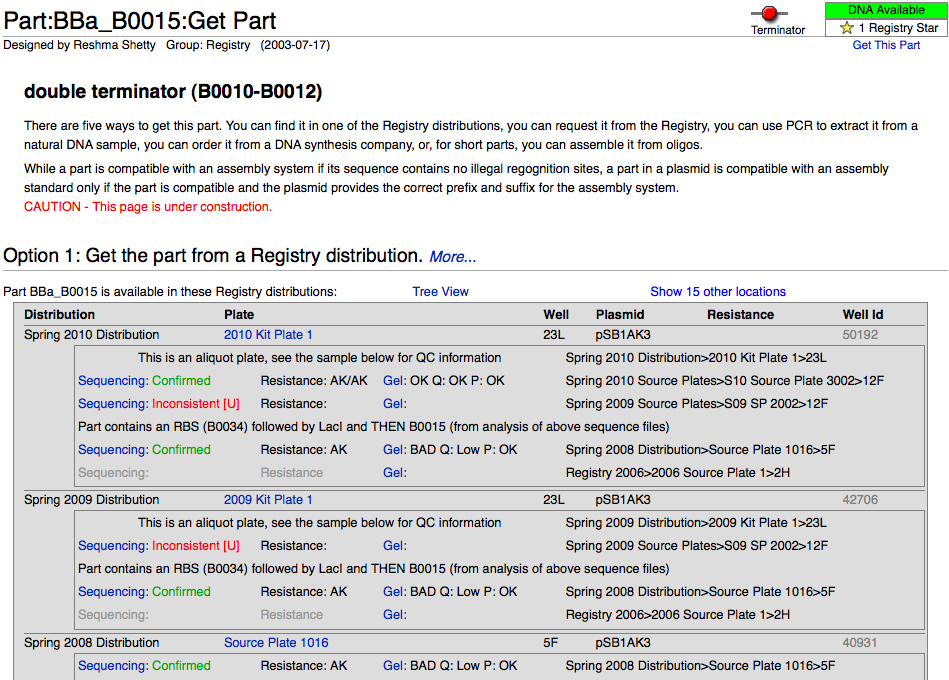
After choosing a particular family of parts in the Catalog, users can select the link identifying that group of parts (i.e., Promoters) to view an explanation of the general structure and function of those parts, as well as information to help better design and document new variations on the parts, and a help section featuring further reading materials. From that introductory page, it is possible to use sorting mechanisms in the Catalog section (for example, categorizing promoters by associated RNA polymerase and by regulatory mechanism) to narrow down the search until arriving at a listing of the desired parts in the registry (for example: Positively regulated E. coli σ70 promoters). These parts have a green box filled by an "A" to the left of the row if they are available. At this point, by selecting the name of the part (Ex: BBa_I1051) it is possible to view further description of the part's features; clicking on the link (Get This Part) at the top right portion of the screen will yield the plate and well location of the part in the Spring 2010 Distribution.
Focused Search
- Go to the Registry.
- In the search box on the upper right, enter the part number that you are interested in (e.g. BBa_B0015) and click GO.
- Click on the Get This Part link at the top right part of the page. Please note that we only have physical DNA for parts whose part status (in the box located just above the Get This Part link) reads Available.
- By following the Get This Part link, you will arrive at a page outlining the location of the part in the parts kit (Spring 2010 distribution), along with QC information and some other ways of obtaining the part if it is not included in this year's parts kit.
Availability
If you decide to do a targeted search for a particular part that you think you might want to use, you need to make sure that the part is actually available. There are many parts in the Registry that people are still working on, or decided not to continue working on anymore, therefore we never received or don't yet have the physical DNA for them. This of course means that the DNA is not available in the Spring 2010 DNA Distribution. The simplest way to tell whether the part is available is to look at the top right of the part's Main Page. If the part is available the top part of the box will be green and say "DNA available."
Ordering a part
We've taken care to create a very well-rounded DNA Distribution this spring, including the high quality parts from previous years, and the some of the 2009 submitted parts. However, should you find the part, or parts, that you require are not available in the distribution but available in the Registry, feel free to send us an email (hq [AT] igem [DOT] org) in order to request it. Just include the part name, the plasmid it's located in, and the source, and we'll send it out to you. For more information please see the Requesting Parts page
Using the online QC resources
For this year's distribution we have carried over our extensive quality control measures conducted in 2008 and 2009, in order to better ensure the accuracy of the parts in the Spring 2010 Distribution. This year every single part has gone through a quality control process which includes AB Test Plates, Growth Plates, Restriction Digests and Gels, and Sequencing.
Here at iGEM, we want everyone to take a look at the results of the quality control measures we've taken this year and previous years, in order to make an informed decision when choosing a part. We've made sure to update the online repository for the Spring 2010 distribution with our quality control results.
The best way to use our quality control information is to use it on a part by part basis. As you design your project, make sure to check every part that you're interested in for its QC data.
- After searching for a part in the registry and arriving at its main page, click on the "Get This Part" link which will take you to the section listing various quality control information for that part and its locations within the registry.
- There are a few different quality control ratings and details to see on this page.
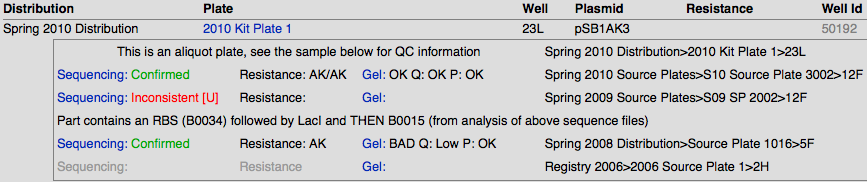
Sequencing Results
The sequences are compared with their target sequence through software, and are given the following qualitative values:
- Confirmed
- Partially Confirmed
- Long Part – we don’t know whether the entire part is confirmed, but the sequence ends are
- Inconsistent
- Bad Sequence – usually caused by low DNA concentration or incorrect primers
- [U] - User Confirmed – we manually reevaluate the inconsistent sequences and look at the trace files to see if a simple shift of the sequence will confirm it
For more in-depth analysis, when reviewing quality control information for a part, just click on the "Sequence" link, right beside the part's Sequence result. Every user can look at all the results we get back from Genewiz: including the trace files, quality scores, and sequence reads.
Gel Results
We did restriction digests on parts using EcoR1 and Pst1 as our restriction enzymes. Afterwards we ran the digests on an e-gel and then imaged the gel using standardized parameters. The gel image was then uploaded to the DNA repository. We evaluated each lane, using a set of qualitative statements for plasmid length (P), plasmid quantity (Q), and insert length (Gel).
- Plasmid Quantity (P): None / LOW / OK / HIGH
- Plasmid Length/Quality (Q): OK / BAD / ???
- Insert Length/Quality (Gel): OK / BAD / ???
To view the gel results for yourself, you can click on the "Gel" link beside the gel result for your part or you can click on the "Gel Images and Results" link at the Source Plate.
Evaluating the Gel Results
On the Gel Images and Results page, you can find the expected length of the part (insert) and the plasmid, as well as the gel image. This allows you to compare the expected lengths of the part and plasmid with their gel bands.
If you find that the gel results for your part do not match with their expected lengths then you may first want to take a look at the part’s Main Page to find out its restriction sites. Some parts and plasmids may have more than one or no EcoR1 and Pst1 cut sites; which will of course differ from the expected banding on the gel. When evaluating the parts we made sure to take these exceptions into consideration; if the gel results matched the band length calculations, the part was described as OK.
Growth Plates and Antibiotic Test Plates
All source plates have images of the pelleted growths before they were sent for miniprepping, and their pelleted growths on 4 antibiotic test plates. In order to view this information for your part, you should know your part's 2010 Source Plate, which you can find on the Get This Part page. From the DNA repository, find the 2010 Source Plate of your choice, and links to the growth and AB test plate images will be at the top.
What do AB test plates tell you?
Antibiotic resistance is measured in the resistance field, which compares the expected antibiotic resistance with the demonstrated antibiotic resistance.
- Whether a part grew up in the AB broth that its plasmid was resistant to.
- Whether the part’s plasmid is resistant to more than one AB.
If there are discrepancies between the plasmid AB description and the AB test, take a look at the gel results, to find out if the plasmid quality (P) was bad.
- If the quality (P) was BAD, the plasmid that the part is in may be incorrect. If so, check the insert length to see if the part is wrong as well.
- If the quality was OK for the plasmid, as well as the insert length, and the sequencing, then it is likely that the AB Test Plate(s) may have had an insufficient antibiotic concentration.