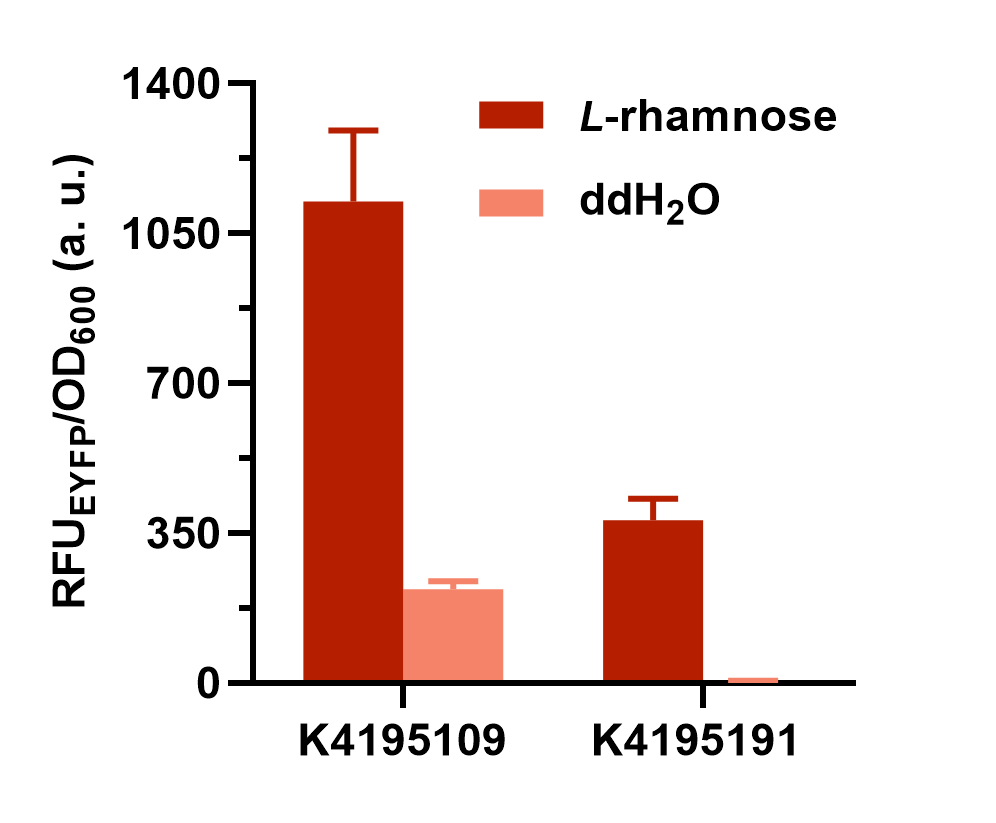Difference between revisions of "Part:BBa K914003"
| Line 59: | Line 59: | ||
[[Image:Image004.jpg|thumb|530px|center|<font size="1">Fitted standard curve of purified Kai A proteins to determine ng of protein from intensity readings in Image J. </font>]] | [[Image:Image004.jpg|thumb|530px|center|<font size="1">Fitted standard curve of purified Kai A proteins to determine ng of protein from intensity readings in Image J. </font>]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K914003 SequenceAndFeatures</partinfo> | <partinfo>BBa_K914003 SequenceAndFeatures</partinfo> | ||
Revision as of 15:05, 12 October 2022
L-rhamnose-inducible promoter (pRha)
L-rhamnose-inducible promoter is capable of high-level recombinant protein expression in the presence of L-rhamnose, it is also tightly regulated in the absence of L-rhamnose by the addition of D-glucose. Read about this biobrick part [http://2012.igem.org/Team:Paris_Bettencourt/Restriction_Enzyme in the context of the project].
Biology of pRha
L-rhamnose is taken up by the RhaT transport system, converted to L-rhamnulose by an isomerase RhaA and then phosphorylated by a kinase RhaB. Subsequently, the resulting rhamnulose-1-phosphate is hydrolyzed by an aldolase RhaD into dihydroxyacetone phosphate, which is metabolized in glycolysis, and L-lactaldehyde. The latter can be oxidized into lactate under aerobic conditions and be reduced into L-1,2-propanediol under unaerobic conditions.
The genes rhaBAD are organized in one operon which is controlled by the rhaPBAD promoter. This promoter is regulated by two activators, RhaS and RhaR, and the corresponding genes belong to one transcription unit which is located in opposite direction of rhaBAD. If L-rhamnose is available, RhaR binds to the rhaPRS promoter and activates the production of RhaR and RhaS. RhaS together with L-rhamnose in turn binds to the rhaPBAD and the rhaPT promoter and activates the transcription of the structural genes. However, for the application of the rhamnose expression system it is not necessary to express the regulatory proteins in larger quantities, because the amounts expressed from the chromosome are sufficient to activate transcription even on multi-copy plasmids. Therefore, only the rhaPBAD promoter has to be cloned upstream of the gene that is to be expressed. Full induction of rhaBAD transcription also requires binding of the CRP-cAMP complex, which is a key regulator of catabolite repression.
The pRha sequence was containing an EcoRI restriction site, so we had to disrupt it in order to use pRha as a biobrick. In order to decide which base pair to modify, we used the [http://microbes.ucsc.edu UCSC Microbial Genome Browser]. We compared the pRha sequence in E.coli and similar species, and identified that the at the position is sometimes replaced by a in some species, so we decided to replace it in a same way. We ordered a gBlock with the pRha sequence having the mutation, and this is the sequence we used and submitted.
Characterization of pRha
Experimental setup
In order to characterize this promoter, we made a construct with a medium RBS (B0032) and an RFP cloned downstream of the pRha, on the pSB3C5 plasmid. We induced the expression of RFP by adding L-Rhamnose. As a negative control, we used cells without the inducer, as well as cells repressed with Glucose.
Results
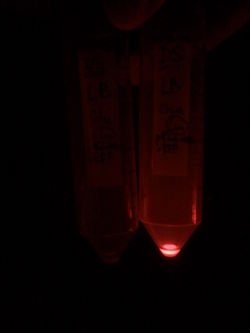
First, by simple observation under a fluorescence viewer, we have seen that the addition of 1% L-Rhamnose leads to a significant expression of RFP after 10hours. The negative controls, where no Rhamnose was added, or when the promoter was repressed by Glucose, did not show any visible fluorescence. Both photos are taken after we centrifuged a culture of NEB Turbo strain with transformed plasmid. For the fluorescent result, the same tubes were photographed under excitation light (540nm), through an emission filter (590nm).
We quantified this result in a plate reader.
Next, we characterized the pRha promoter using a plate reader. We used different concentrations of L-Rhamnose (0.05%, 0.1%, 0.2%, 0.5% and 1%) and observed the resulting fluorescence over time. As negative controls, we used the non-induced cells, as well as cells repressed by 1% Glucose.
As we can see from the graph above, pRha promoter works as expected and it could be well tuned by concentration of L-rhamnose.
The UChicago 2015 iGEM team also used this part in their project and did some characterization of the promoter, using Western blot for their gene of interest, providing an improvement on the characterization accomplished by the 2012 Paris-Bettencourt team which used a plate-reader to measure fluorescence. In contrast to the low copy plasmid used by the Paris Bettencourt team, they characterized the promoter when expressed in a high copy plasmid. Accordingly, they discovered that despite following a similar protocol (10 hours of induction), significantly less rhamnose was required for the induction of the promoter in the high copy number plasmid.
The UChicago team also verified that the pRha promoter is quite tight and has a relatively gradual response curve, characteristics that makes the promoter especially well-suited for assaying a wide range of concentrations of a protein of interest.
The densitometry results from Western blots indicate the tightness of pRha. At 0% L-rhamnose, there is significantly less KaiA expressed compared to KaiB and KaiC on a separate constitutively expressed promoter. This supports our ability to tune KaiA concentrations to optimize oscillations.
The gradient curve developed portrays the ability to fine tune the concentrations of KaiA. The data in this graph was corrected using ng from protein standards (below)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team GreatBay_SCIE 2022: improvement on the expression level of pRha promoter
From pRha to pRha-RiboJ
The part uses a rhamnose-inducible promoter system to achieve high level expression of GOI with the fusion of a ribozyme at the junction of the promoter and its downstream sequence.
The part is an improved version based on existing pRha part (BBa_K914003), which is characterized by its high protein expression ability. The part was later modified in the construction of pRha-riboJ-Neae-Nb to enhance expression of Nb which enables cellulosome complex to be displayed on surface of E.coli.
Ribozyme (ribonucleic acid enzymes) are RNA molecules with a variety of different functions including RNA splicing in gene expression. A synthetic self-cleaving ribozyme called RiboJ was used in this circumstance to increase the RFP expression.
RiboJ is a 75 nucleotide sequence consisted of a satellite RNA of tobacco ringspot virus derived ribozyme and a 23 nucleotide hairpin [1]. During post-transcriptional processing of mRNA, RiboJ self-cleaves to eliminate its upstream sequence and remove the promoter-associated RNA leader. Consequently, only the uncleaved hairpin sequence is presented before the RBS and gene of interest. This genetic insulation would increase gene expression level at both RNA and protein level. Gene insulation by RiboJ increases gene expression via stabilizing mRNA and reducing the degradation of the transcript, resulting in higher transcript abundance [1]. The greater mRNA abundance is then amplified by translation, elevating the production of target protein.
Characterization
The Expression of pRha-riboJ-RFP and pRha-RFP
The ability for ribozyme RiboJ to increase expression level of gene of interest downstream was tested with the composite part pRha-riboJ-RFP (Fig.1A), which is an improvement of the pRha-RFP part.
The RFP production of both pRha-riboJ-RFP group and pRha-RFP control group was quantified by fluorescence intensity and riboJ group displays significant increment in level of protein expression compared to corresponding construct without riboJ (Fig.1C and Fig.1D).
The Expression of pRha-riboJ-Neae-Nb
The results of pRha-riboJ-RFP expression successfully verified the effect of RiboJ gene insulation on increasing gene expression. We then used the part as a vector and substitute RFP sequence with coding sequence Neae-Nb (Fig.1A), which is part of the Nb-Ag3 surface display system. We cultured E. coli. BL21 with this construct, induced its expression and collected the cells. SDS-page was performed, indicating that Neae-Nb was successfully expressed (Fig.1B).

Verification of protein-protein interaction
In order to verify the natural function of Neae-Nb-Ag3 surface display system, we constructed E. coli expression vector for Ag3 domain ligated with eforRed domain to visualize the protein-protein interaction (Fig.2B). The Ag3-eforRED construct was cultured for IPTG-inducible expression. Desired proteins were identified in whole cell and supernatant samples as shown by SDS-page analysis (Fig.2C). Intact E. coli cells expressing Neae-Nb on their surface was mixed with eforRed-Ag3 supernatant. In contrast to the control group only with Neae-Nb, red fluorescence was observed in Neae-Nb-Ag3 mixture under blue-light condition, providing evidence for Nb-Ag3 interaction (Fig.2D).

Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 91
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 91
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 91
- 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 91
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 91
- 1000COMPATIBLE WITH RFC[1000]
References
1.Clifton, Kalen P et al. “The genetic insulator RiboJ increases expression of insulated genes.” Journal of biological engineering vol. 12 23. 29 Oct. 2018, doi:10.1186/s13036-018-0115-6
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
Team Aix-Marseille 2020 : Bibliographic research to ameliorate the PrhaBAD biobrick efficiency
RhaS
When using this promoter as a biosensor, it is used with only one gene encoding a fluorescent protein (rfp, gfp).This is because the Paris_Bettencourt 2012 team thought that it was sufficient to have a good expression by rhamnose. But we learned that if we add in the plasmid a gene that encodes constitutively for RhaS, we could have a better fluorescence.1 We could have decoupled the RhaS protein to the RhaR protein, thus suppressing the activator dependency.1 More, it allows us to import this biobrick in other bacteria than E.coli which don’t have rhaS gene in their chromosome. This modification has already been tested in cyanobacteria which is a clade within it’s difficult to find a good inducible promoter system, which is no toxic, with a linear expression.2 A study shows that Synechocystis sp, a cyanobacteria, has efficiently produced protein without either an E.coli sigma factor or a protein homologous to RhaS.2
RhaB/RhaT
However, the efficiency of the PrhaBAD system can be increased yet.
We know for exemple that the rhamnose is metabolized by E.coli, so the quantity in the cell decreases with time which involves that fluorescent signal being transient in function of time.
So for the chassis, we can use a bacteria where the chromosomic rhaB is inactivated by introduction of a shift mutation in rhaB.3 Thus, L-rhamnose isn’t metabolized anymore.3 This modification allows to have a signal that isn’t transient anymore for the protein production but neither dose dependant.3 The rhaB mutation alone allows us to have a response « all or nothing » independent of catabolism.3
Another point to ameliorate is the false dose-dependant effect of the PrhaBAD biobrick.3 In fact, some studies show that effect is due to the consumption of L-rhamnose by metabolism and not due to the regulation of production yield.3
Thus, to increase the accuracy of fluorescence measures, we can mutate rhaB and also suppress the active transport of rhamnose done by RhaT by suppressing its gene.3 Thus the signal will be only dependent on the passive diffusion of rhamnose and so dependent on the media concentration of rhamnose.3 These two mutations allow us to have a real accurate dose-dependant effect in fonction of the concentration of rhamnose in media and independent of the catabolism.3
These modifications have the advantage to not decrease the growth of the bacteria.3 But they allow overall to know exactly which concentration of inducer is optimal to produce a chosen protein efficiently.3
The double mutation described here allows also to have a better yield of secretory protein production which can have a saturation at the level of their biogenesis pathway.3
New inducer : L-mannose
This promoter can be induced by an analogue of L-rhamnose, the L-mannose, which has a close effect but which isn’t metabolised.1 The article cited earlier tells us that the L-mannose could be a perfect analogue.1 Our system described previously is sometimes even more sensitive for it than for L-rhamnose.1 It also has the same linear response as with the L-rhamnose.1 So we have a response that continues in time and is more powerful.1 That surely because L-mannose isn’t well metabolized by E.coli and because it is recognized by RhaS.1 We also have an unimodal induction like with L-rhamnose which is useful to produce the same concentration of protein in different cells, except for low inducer concentration where the expression is bimodal.1 This bimodality at low concentration is also encountered with the Plac/IPTG inducible system which is hugely used, so it isn’t really a problem.1 It can be resolved by the use of another analogue, the L-lyxose, less efficient than the L-mannose but unimodal at low concentration.1 More, these two analogues are easily found in stores.1 The problem can however be the price of L-mannose, relatively expensive.1
- Kelly et al, 2016 Synthetic Chemical Inducers and Genetic Decoupling Enable Orthogonal Control of the rhaBAD Promoter
- Kelly et al, 2018 A Rhamnose-Inducible System for Precise and Temporal Control of Gene Expression in Cyanobacteria
- Hjelm, 2017 Tailoring Escherichia coli for the L‑Rhamnose PBAD Promoter-Based Production of Membrane and Secretory Proteins
Team XMU-China 2022: improvement the behavior of pRha
Background
pRha (BBa_K914003) is a strictly regulated promoter, which was firstly registered in 2013 and has been widely used in stringently expression of GOI (gene of interest) (1-3) . RhaS is a transcriptional activator of pRha, which will sense L-rhamnose and allosterically bind pRha to activate the transcription. In addition, previous work has showed that another inducer, L-rhamnose, could activate pRha with a better behavior (4) , thus we also included the L -mannose-induced characterization of pRha, compared with L-rhamnose.
Design
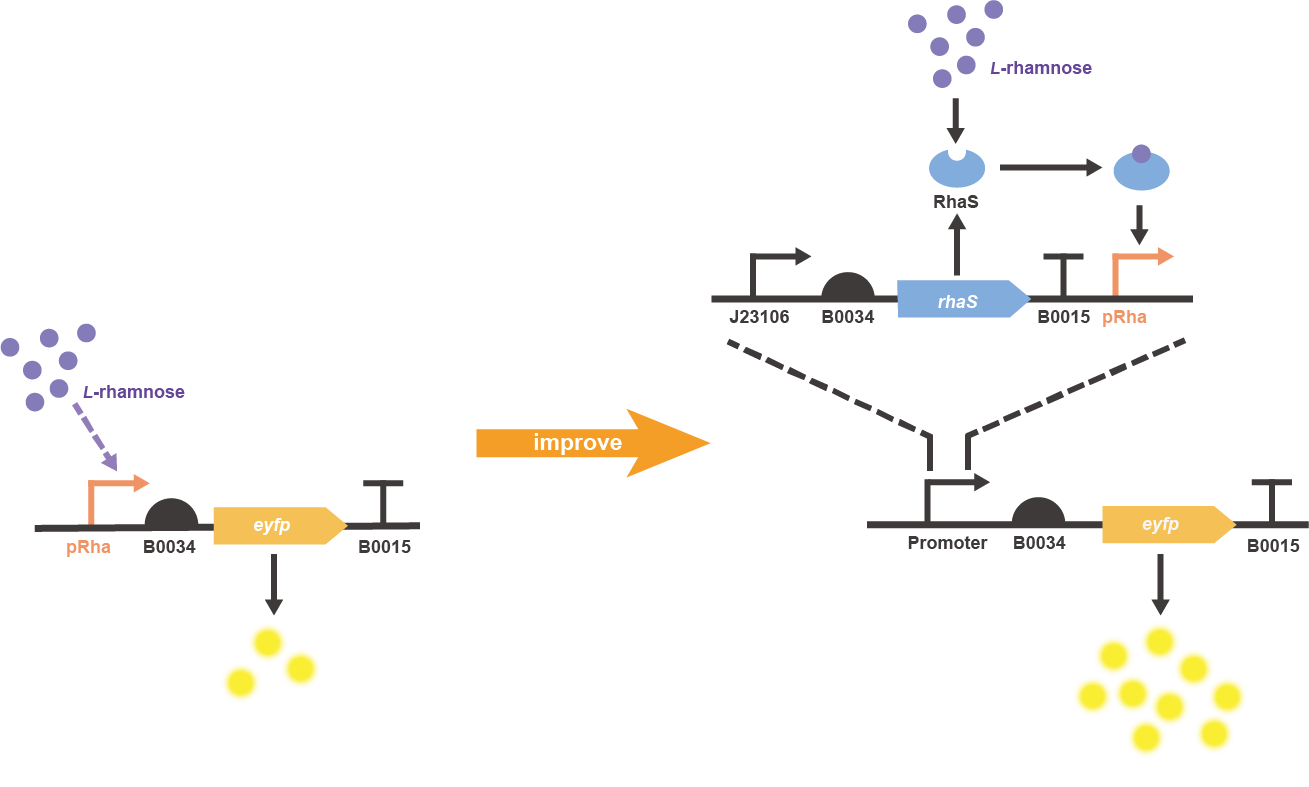
In order to improve the induced expression level of L-rhamnose-inducible promoter BBa_K914003, an extra copy of rhaS gene was constitutively expressed and placed upstream the pRha, in which the eyfp gene (BBa_E0030) was chosen as the reporter to finally construct the two characterization circuits of original and improved parts (Fig. 1).
Fig. 1 Gene circuit illustration for improved pRha promoter system.
Result
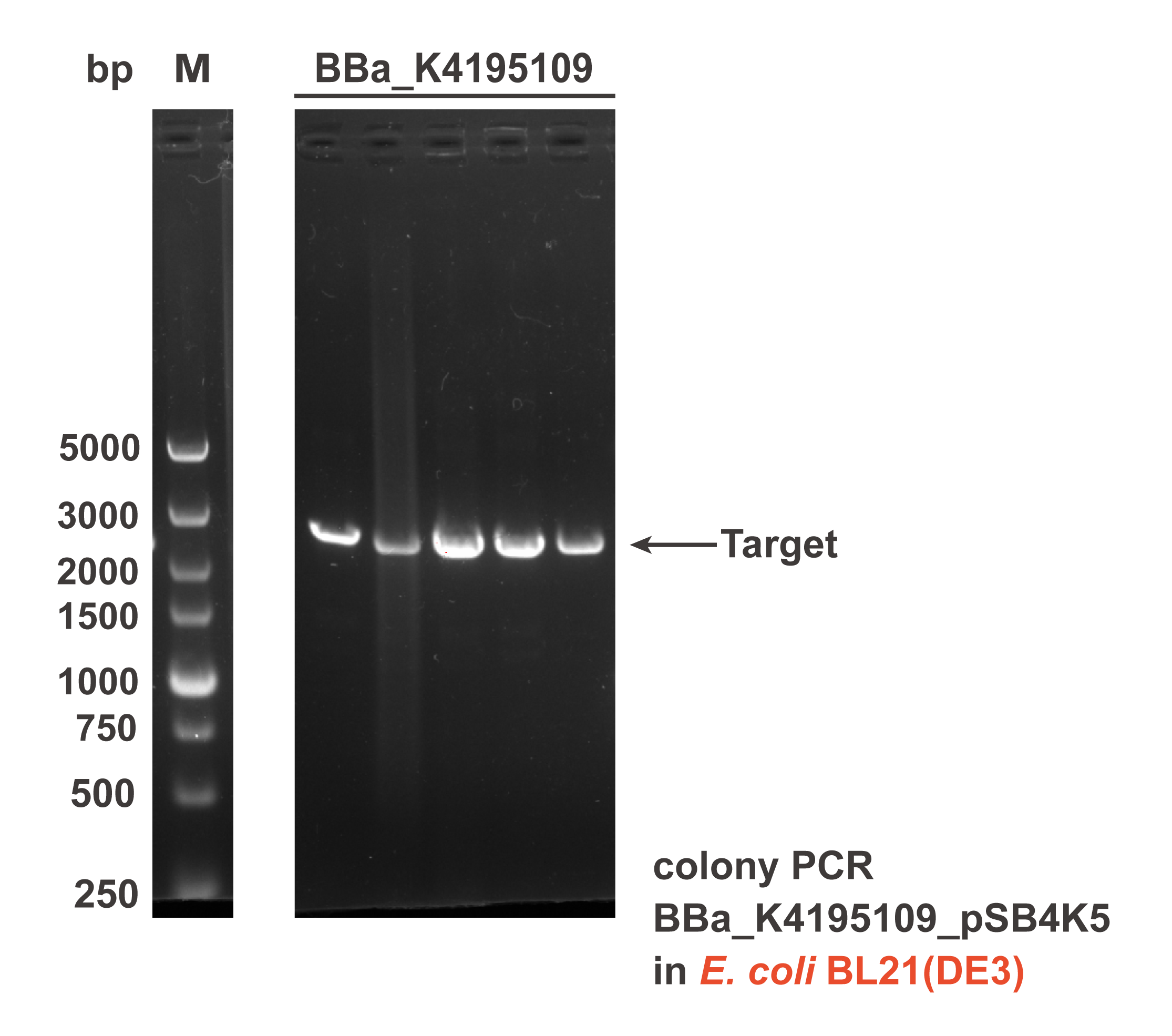
Different sub parts of the two circuits were assembled into pSB4K5 plasmid backbone using Gibson assembly to get the composite part BBa_K4195109 and BBa_K4195191. The Gibson assembly reaction mixture was transformed into E. coli DH5α and E. coli BL21(DE3), then the correct colonies were confirmed by colony PCR (Fig. 2) and sequencing.
Fig. 2 The result of colony PCR. Then, these colonies harboring the corrected plasmid were cultivated and induced to measure the expression level of EYFP (eyfp). Both the composite parts were induced under L-rhamnose (final concentration was 0.1 mg/mL) and the sterile water of same volume was added as the control. The fluorescence intensity (λEx = 507 nm, λEm = 532 nm) and OD600 was measured in 2 h after induction. The relative fluorescent unit (RFU) normalized to OD600, RFUEYFP/OD600, was calculated to represent the expression level of EYFP in an average cell. We can obviously find that the gene circuit with rhaS showed higher level of fluorescence signal, even though a little bit of leakage expression was observed (Fig. 3).
Fig. 3 The comparison of normalized fluorescence intensity between BBa_K4195109 and BBa_K4195191. What’s more, we also set a series of concentration gradients to compare different inducers’ effects on the improved L-rhamnose-inducible promoter. Learn more specific and detailed information in the parts page of this composite part (BBa_K4195109).
Reference
1. S. K. Kim, D. H. Lee, O. C. Kim, J. F. Kim, S. H. Yoon, Tunable Control of an Escherichia coli Expression System for the Overproduction of Membrane Proteins by Titrated Expression of a Mutant lac Repressor. ACS Synth. Biol. 6, 1766-1773 (2017).
2. A. Haldimann, L. L. Daniels, B. L. Wanner, Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 180, 1277-1286 (1998).
3. Q. Li et al., A modified pCas/pTargetF system for CRISPR-Cas9-assisted genome editing in Escherichia coli. Acta Biochim. Biophys. Sin. 53, 620-627 (2021).
4. C. L. Kelly et al., Synthetic Chemical Inducers and Genetic Decoupling Enable Orthogonal Control of the rhaBAD Promoter. ACS Synth. Biol. 5, 1136-1145 (2016).