Difference between revisions of "Part:BBa K4247009"
(→Characterization of Mfp151_SnoopCatcher =) |
|||
| Line 67: | Line 67: | ||
For more information about this protein, go to <partinfo>BBa_K4247025</partinfo>. | For more information about this protein, go to <partinfo>BBa_K4247025</partinfo>. | ||
| − | == Characterization of Mfp151_SnoopCatcher | + | == Characterization of Mfp151_SnoopCatcher == |
For more information about this protein, go to <partinfo>BBa_K4247021</partinfo>. | For more information about this protein, go to <partinfo>BBa_K4247021</partinfo>. | ||
| + | |||
| + | === Addition of SnoopCatcher to Mfp151 === | ||
| + | |||
| + | '''Aim - '''The tyrosine residues in the Mfp151 protein undergo post-translational modifications (PTMs) to become 3,4-dihydroxyphenyl-alanine (Dopa) which provides the Mfp151 proteins with their adhesion properties. Since E.coli is not capable of performing PTMs, this can be overcome by co-expressing another plasmid producing tyrosinase to modify tyrosine residues to Dopa in vivo. Mfp151 and Mfp151_Snoopcatcher were expressed. Further, they were also co-expressed with tyrosinase to facilitate the post-translational modification of tyrosine to Dopa to make the proteins adhesive. | ||
| + | |||
| + | '''Results - '''In the SDS-gel, it is difficult to visualise the Mfp151 protein. So, a western blot was done on the above SDS-gel. | ||
| + | |||
| + | |||
| + | [[File:Mfp13.png]] | ||
| + | |||
| + | |||
| + | [[File:Mfp14.png]] | ||
| + | |||
| + | |||
| + | '''Conclusion - ''' Thus, we can see the expression of Mfp151 (25kDa), Mfp151_Snoopcatcher (37kDa) along with tyrosinase (30kDa) in the co-expression cultures. We cannot see orf438 (cofactor) because it ran out of the gel. In the next experiment however it's visible. | ||
| + | |||
| + | === Protein purification by IMAC === | ||
| + | |||
| + | '''Aim - '''To purify the protein by IMAC (immobilized metal ion chromatography) using Ni-NTA resin. | ||
| + | |||
| + | '''Results - '''Mini columns were loaded with Ni-NTA resin and the soluble fraction of the lysate was added to the columns. Then, the columns were washed twice and eluted to obtain the purified protein. An SDS-gel was run with the different purification fractions and a western blot was done on the gel. It is clear that most of the protein was lost in the flowthrough and washes and nothing was eluted, showing that the proteins did not bind to the Ni-NTA column at all. | ||
| + | |||
| + | Lysis buffer - 10mM Tris-Cl, 100mM NaH2PO4, 8M Urea, pH 8 | ||
| + | |||
| + | Wash buffer - 10mM Tris-Cl, 100mM NaH2PO4, 8M Urea, pH 6 | ||
| + | |||
| + | Elution buffer 1 - 10mM Tris-Cl, 100mM NaH2PO4, 8M Urea, pH 4.5 | ||
| + | |||
| + | Dialysis buffer - distilled water | ||
| + | |||
| + | [[File:mfpcpuri.jpeg]] | ||
| + | |||
| + | '''Conclusion - '''Although a considerable amount of the protein was lost in the flowthrough and washes, it is clear that the protein is eluted with high purity. | ||
| + | |||
| + | ===Co-transformation with tyrosinase and cofactor=== | ||
| + | |||
| + | '''Aim - ''' To show that the co-transformation of the pET24 (+) vector containing the mfp151 (BBa_K4247018-BBa_K4247021) and the pRSET A vector containing tyrosinase (BBa_K4247023) and orf438 (tyr-cofactor) (BBa_K4247022) works. | ||
| + | |||
| + | '''Result - ''' SDS and Western Blot was done on the purification fractions obtained from Ni-NTA purification of the protein from BL21(DE3) cells induced with 0.1 mM IPTG overnight. As it can be observed, in the SDS, tyrosinase (31.56 KDa) and orf438-cofactor- (16.48 KDa) are being produced. Since Mfp151 does not have any tryptophan residues, it is not possible to visualise Mfp151 proteins in an SDS-gel and hence, a western blot is needed. | ||
| + | |||
| + | [[File: sdst.jpeg|700px|]] | ||
| + | |||
| + | So, a western blot was done on the above SDS-gel to confirm that the proteins we see are indeed the minispidroin proteins. Since the proteins were expressed with a 6x His-tag, we used mouse anti-hexa his primary antibodies and goat anti-mouse HRP-conjugated secondary antibodies for the western blot. | ||
| + | |||
| + | [[File: wbt.jpeg|700px|]] | ||
| + | |||
| + | '''Conclusion - '''As seen in the Western Blot, we lost proteins in the flowthrough and washes. However, it still proves we managed to produce mfp151 and mfp151_SnoopCatcher in co-transformation with tyrosinase and orf438. The cofactor is clearly visible in the SDS-gel but not so clear in the Western Blot, possibly because the 6x HisTag is not well exposed. | ||
| + | |||
| + | === NBT assay === | ||
| + | |||
| + | '''Aim - '''To qualitatively evaluate the amount of tyrosine residues modified into DOPA-tyrosine and understand whether the co-expression system of mfp151 + orf438-tyrosinase operon made mfp151 more adhesive. The final goal was to test whether we could increase the interactions between mfp151 and minispidroins. | ||
| + | |||
| + | '''Results - '''We performed an NBT assay (Nitro-Blue Tetrazolium), where the purified and dialysed proteins (mfp151, mfp151 + tyrosinase coexpression, mfp151_SnoopCatcher and mfp151_SnoopCatcher + tyrosinase coexpression) were treated with a 0.6 mg/mL NBT solution and washed with a 0.16 M potassium glycinate solution. | ||
| + | |||
| + | [[File: Nbtdopa.jpeg]] | ||
| + | |||
| + | '''Conclusion - '''Mfp151 is clearly the most stained protein, indicating that the co-expression system did not work effectively. This could be due to multiple reasons, such as a bias from other metal binding proteins that were not removed effectively during IMAC purification. After this result, we decided to implement a more precise protein conjugation tool as an alternative to making the Mfp151 more sticky. | ||
| + | |||
| + | === Validation of SnoopCatcher's function === | ||
| + | |||
| + | '''Aim - '''To demonstrate the spontaneous isopeptide bond formation between our minispidroin protein displaying the SnoopTag and Mfp151_SnoopCatcher. | ||
| + | |||
| + | '''Results - '''The proteins were allowed to interact with each other by mixing equal amounts of both proteins. The reaction was carried out at 25°C for 40 minutes and the pH of the reaction was maintained at 8 using pH adjusted PBS. After 40 minutes, the reaction was stopped by adding SDS sample buffer. Then, we performed an SDS-PAGE and a Western blot to visualize the results. We would expect to see a band whose molecular weight is the sum of the molecular weights of both proteins to indicate that both proteins are bound to each via an isopeptide bond between the SnoopTag and the SnoopCatcher. We see such a band only in the reaction where Mfp151_SnoopCatcher and Minispidroin 2rep with a SnoopTag were allowed to interact. The fact that we don't see this band in any of the controls confirms that the proteins were able to bind to each other solely due to the SnoopTag-Catcher system. | ||
| + | |||
| + | [[File:mfpcpoc.jpeg]] | ||
| + | |||
| + | '''Conclsuion - '''The SnoopCatcher enabled the Mfp151_SnoopCatcher to bind to Minispidroin 2rep with a SnoopTag and hence, the function of the SnoopTag-Catcher system was validated. | ||
Revision as of 15:05, 12 October 2022
Contents
SnoopCatcher
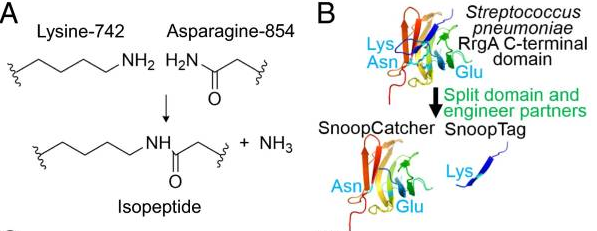
This part codes for SnoopCatcher, a short peptide that together with the basic part BBa_K4247008 (SnoopTag) enables to link proteins through the formation of a covalent bond between Tag and Catcher. It has been used extensively to form polyproteins in a spontaneous reaction (but can be supported by SnoopLigase).
Usage and Biology
The Tag-Catcher system is a protein conjugation tool that enables the formation of an irreversible isopeptide bond between these two components. A covalent peptide/protein pair was developed to enable spontaneous isopeptide bond formation between peptide tags. This system was developed based on a Gram-positive surface protein, the pilus adhesin RrgA of S. pneumoniae. The D4 domain of this protein is stabilised by an isopeptide forming between a lysine (K742) and an asparagine (N854). This domain was split into a scaffold protein called SnoopCatcher and a 12-residue peptide termed SnoopTag, which can spontaneously form a covalent isopeptide bond upon mixing. The initiation, extension, and release steps use mild conditions, independent of redox state, and therefore should be applicable to a wide range of proteins according to the original paper.
Characterization of mCherry_SnoopCatcher
Addition of SnoopCatcher to mCherry
Aim - To attach the SnoopCatcher sequence (Part BBa_K4247009) to the original mCherry part (Part BBa_J06504).
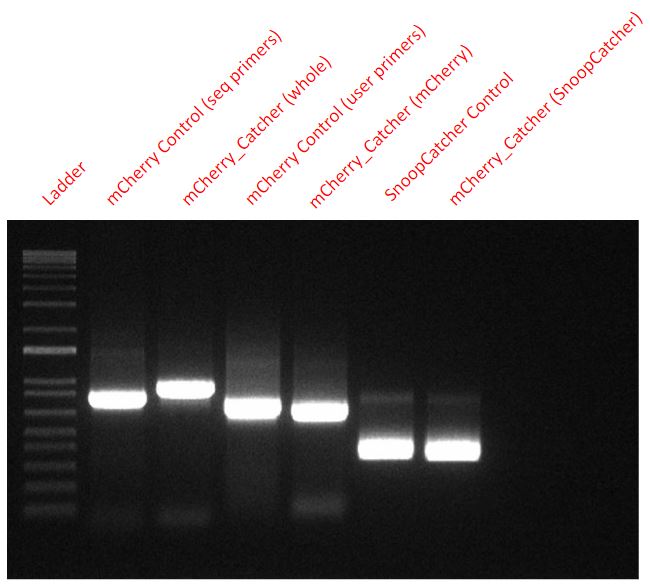
Results - We designed 6 primers with Uracil to perform a USER cloning reaction. Two primers to amplify the mCherry fragment while swapping the 6-His tag’s location from the C-terminus to the N-terminus, two for amplifying the backbone of the expression vector (pBAD) and two for amplifying the SnoopCatcher fragment from a part we already produced (BBa_K4247021 - Mfp151_SnoopCatcher). We verified the success of the cloning via PCR and sequencing.
From the final USER cloned plasmid coding for the mCherry_SnoopCtacher, we amplified mCherry, SnoopCatcher and the whole mCherry_SnoopCatcher fragment via PCR to confirm that our USER cloning worked. We also used mCherry and SnoopCatcher fragments as controls.
From a plasmid expressing just the mCherry, we used two sets of primers (seq primers and USER primers) to amplify the mCherry fragment as a control for mCherry. However, the seq primers also amplified some base pairs that were upstream and downstream and hence, one of the control fragments - mCherry Control (seq primers) - seems slightly higher than the other mCherry control fragment.
Conclusion - Hence, it is clear that our USER cloning was successful in adding the SnoopCatcher to the mCherry fragment as seen in the PCR.
Protein expression of mCherry_SnoopCatcher
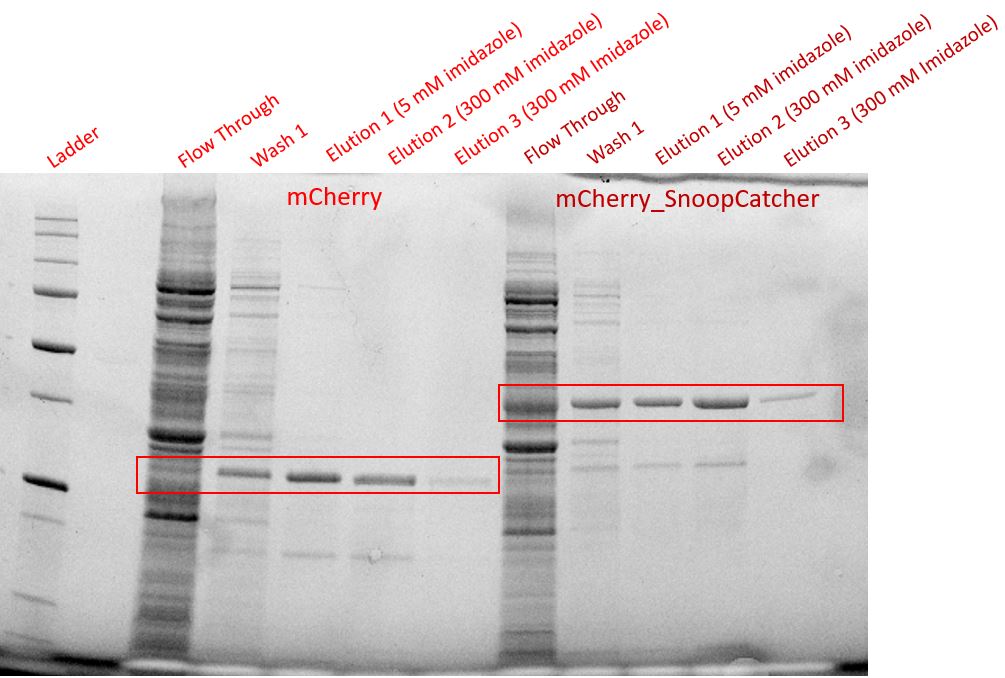
Aim - To express and purify mCherry_SnoopCatcher.
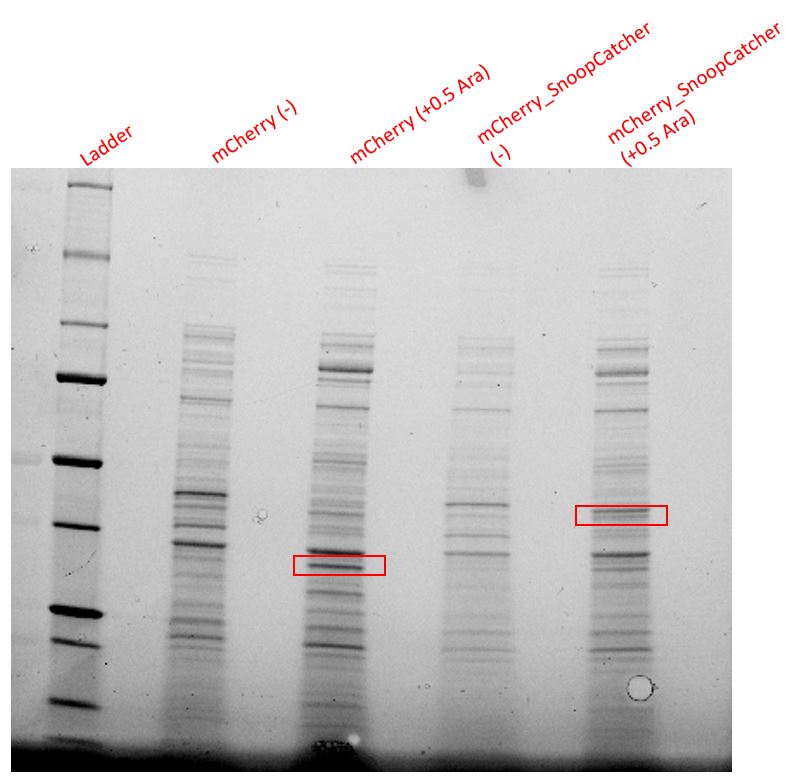
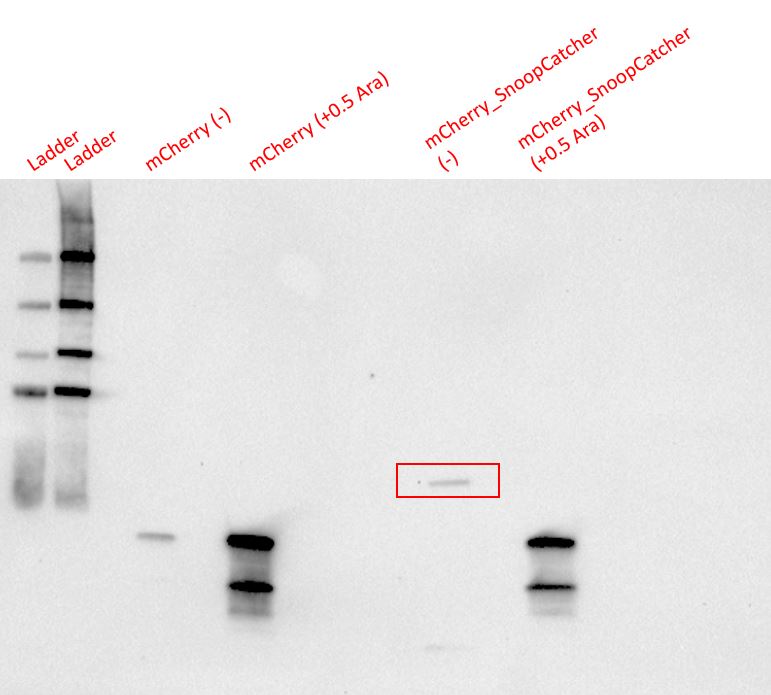
Results - We expressed the mCherry_SnoopCatcher and mCherry in E.coli BL21(DE3) and induced protein expression using 0.5% arabinose overnight. The next day, we obtained a bright red E.coli culture for both proteins. Then, we performed SDS-PAGE and a Western blot to check for protein expression. It was clear that both proteins were expressed since we found bands of appropriate size only in the induced lanes.
We then performed IMAC using Ni-NTA resin since the protein was expressed with a 6x His-tag and successfully purified mCherry_SnoopCatcher as well as mCherry. Then, we performed SDS-PAGE and a Western blot on the purification fractions of both proteins which confirmed that both mCherry_SnoopCatcher (40.89 KDa) and mCherry (26.7 KDa) were expressed since we found bands at the expected molecular weights. Further, we noticed that mCherry has a leaky expression (see mCherry control in the western blot) and that mCherry_Catcher does not stain as well as mCherry. The purification was not as efficient since an important portion of the protein was lost in the washes, probably due to saturation of the beads. However, we only needed small amounts of proteins for our assays.
Conclusion - Thus, mCherry and mCherry_SnoopCatcher were successfully expressed and purified.
Effect of the SnoopCtacher on mCherry’s fluorescence
Aim - To ensure the addition of the SnoopCatcher would not impair the main characteristic of mCherry, its fluorescence.
Results - We noticed that mCherry_SnoopCatcher also emits fluorescence in the red wavelengths. It was clear from the lysates that mCherry_SnoopCatcher fluoresces just as much, if not more than the original mCherry, considering that the lysate of mCherry comes from a 0.5L culture while the lysate of mCherry-SnoopCatcher comes from a 150mL culture. This leads us to believe that the addition of SnoopCatcher might improve the protein expression of mCherry or might even improve the fluorescence property of mCherry. However, these results need to be validated since we could not quantify and compare the protein yields or the fluorescence intensity due to time constraints.
Conclusion - Hence, the addition of SnoopCatcher does not affect the properties of mCherry.
Validation of SnoopCatcher’s function
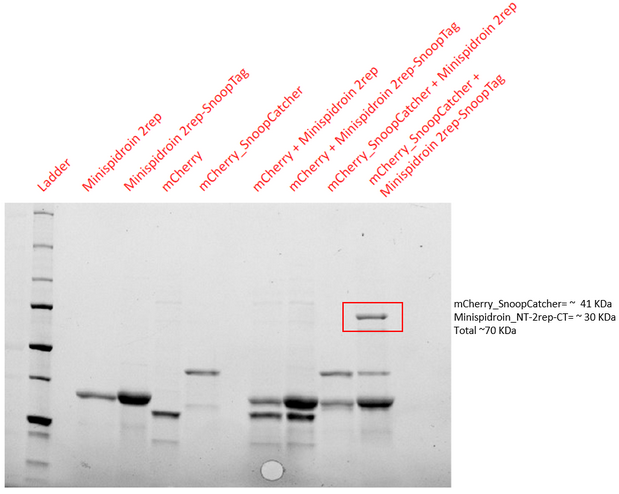
Aim - To demonstrate the spontaneous isopeptide bond formation between our minispidroin protein displaying the SnoopTag and mCherry_SnoopCatcher.
Results - The proteins were allowed to interact with each other by mixing equal amounts of both proteins. The reaction was carried out at 25°C for 40 minutes and the pH of the reaction was maintained at 8 using pH adjusted PBS. After 40 minutes, the reaction was stopped by adding SDS sample buffer. Then, we performed an SDS-PAGE and a Western blot to visualize the results. We would expect to see a band whose molecular weight is the sum of the molecular weights of both proteins to indicate that both proteins are bound to each via an isopeptide bond between the SnoopTag and the SnoopCatcher. We see such a band only in the reaction where mCherry_SnoopCatcher and Minispidroin 2rep with a SnoopTag were allowed to interact. The fact that we don't see this band in any of the controls confirms that the proteins were able to bind to each other solely due to the SnoopTag-Catcher system.
Conclusion - The SnoopCatcher enabled the mCherry_SnoopCatcher to bind to Minispidroin 2rep with a SnoopTag and hence, the function of the SnoopTag-Catcher system was validated.
For more information about this protein, go to BBa_K4247025.
Characterization of Mfp151_SnoopCatcher
For more information about this protein, go to BBa_K4247021.
Addition of SnoopCatcher to Mfp151
Aim - The tyrosine residues in the Mfp151 protein undergo post-translational modifications (PTMs) to become 3,4-dihydroxyphenyl-alanine (Dopa) which provides the Mfp151 proteins with their adhesion properties. Since E.coli is not capable of performing PTMs, this can be overcome by co-expressing another plasmid producing tyrosinase to modify tyrosine residues to Dopa in vivo. Mfp151 and Mfp151_Snoopcatcher were expressed. Further, they were also co-expressed with tyrosinase to facilitate the post-translational modification of tyrosine to Dopa to make the proteins adhesive.
Results - In the SDS-gel, it is difficult to visualise the Mfp151 protein. So, a western blot was done on the above SDS-gel.
Conclusion - Thus, we can see the expression of Mfp151 (25kDa), Mfp151_Snoopcatcher (37kDa) along with tyrosinase (30kDa) in the co-expression cultures. We cannot see orf438 (cofactor) because it ran out of the gel. In the next experiment however it's visible.
Protein purification by IMAC
Aim - To purify the protein by IMAC (immobilized metal ion chromatography) using Ni-NTA resin.
Results - Mini columns were loaded with Ni-NTA resin and the soluble fraction of the lysate was added to the columns. Then, the columns were washed twice and eluted to obtain the purified protein. An SDS-gel was run with the different purification fractions and a western blot was done on the gel. It is clear that most of the protein was lost in the flowthrough and washes and nothing was eluted, showing that the proteins did not bind to the Ni-NTA column at all.
Lysis buffer - 10mM Tris-Cl, 100mM NaH2PO4, 8M Urea, pH 8
Wash buffer - 10mM Tris-Cl, 100mM NaH2PO4, 8M Urea, pH 6
Elution buffer 1 - 10mM Tris-Cl, 100mM NaH2PO4, 8M Urea, pH 4.5
Dialysis buffer - distilled water
Conclusion - Although a considerable amount of the protein was lost in the flowthrough and washes, it is clear that the protein is eluted with high purity.
Co-transformation with tyrosinase and cofactor
Aim - To show that the co-transformation of the pET24 (+) vector containing the mfp151 (BBa_K4247018-BBa_K4247021) and the pRSET A vector containing tyrosinase (BBa_K4247023) and orf438 (tyr-cofactor) (BBa_K4247022) works.
Result - SDS and Western Blot was done on the purification fractions obtained from Ni-NTA purification of the protein from BL21(DE3) cells induced with 0.1 mM IPTG overnight. As it can be observed, in the SDS, tyrosinase (31.56 KDa) and orf438-cofactor- (16.48 KDa) are being produced. Since Mfp151 does not have any tryptophan residues, it is not possible to visualise Mfp151 proteins in an SDS-gel and hence, a western blot is needed.
So, a western blot was done on the above SDS-gel to confirm that the proteins we see are indeed the minispidroin proteins. Since the proteins were expressed with a 6x His-tag, we used mouse anti-hexa his primary antibodies and goat anti-mouse HRP-conjugated secondary antibodies for the western blot.
Conclusion - As seen in the Western Blot, we lost proteins in the flowthrough and washes. However, it still proves we managed to produce mfp151 and mfp151_SnoopCatcher in co-transformation with tyrosinase and orf438. The cofactor is clearly visible in the SDS-gel but not so clear in the Western Blot, possibly because the 6x HisTag is not well exposed.
NBT assay
Aim - To qualitatively evaluate the amount of tyrosine residues modified into DOPA-tyrosine and understand whether the co-expression system of mfp151 + orf438-tyrosinase operon made mfp151 more adhesive. The final goal was to test whether we could increase the interactions between mfp151 and minispidroins.
Results - We performed an NBT assay (Nitro-Blue Tetrazolium), where the purified and dialysed proteins (mfp151, mfp151 + tyrosinase coexpression, mfp151_SnoopCatcher and mfp151_SnoopCatcher + tyrosinase coexpression) were treated with a 0.6 mg/mL NBT solution and washed with a 0.16 M potassium glycinate solution.
Conclusion - Mfp151 is clearly the most stained protein, indicating that the co-expression system did not work effectively. This could be due to multiple reasons, such as a bias from other metal binding proteins that were not removed effectively during IMAC purification. After this result, we decided to implement a more precise protein conjugation tool as an alternative to making the Mfp151 more sticky.
Validation of SnoopCatcher's function
Aim - To demonstrate the spontaneous isopeptide bond formation between our minispidroin protein displaying the SnoopTag and Mfp151_SnoopCatcher.
Results - The proteins were allowed to interact with each other by mixing equal amounts of both proteins. The reaction was carried out at 25°C for 40 minutes and the pH of the reaction was maintained at 8 using pH adjusted PBS. After 40 minutes, the reaction was stopped by adding SDS sample buffer. Then, we performed an SDS-PAGE and a Western blot to visualize the results. We would expect to see a band whose molecular weight is the sum of the molecular weights of both proteins to indicate that both proteins are bound to each via an isopeptide bond between the SnoopTag and the SnoopCatcher. We see such a band only in the reaction where Mfp151_SnoopCatcher and Minispidroin 2rep with a SnoopTag were allowed to interact. The fact that we don't see this band in any of the controls confirms that the proteins were able to bind to each other solely due to the SnoopTag-Catcher system.
Conclsuion - The SnoopCatcher enabled the Mfp151_SnoopCatcher to bind to Minispidroin 2rep with a SnoopTag and hence, the function of the SnoopTag-Catcher system was validated.