Difference between revisions of "Part:BBa K1162006"
Yuanhui Sun (Talk | contribs) (→HZAU-China 2021) |
Yuanhui Sun (Talk | contribs) (→Design) |
||
| Line 104: | Line 104: | ||
===Design=== | ===Design=== | ||
| + | <p>In order to fit the project and guide the experiment, our research decided to depict the characterization of LL-37 protein- an antimicrobial peptide.</p> | ||
| + | <p>The plasmid we designed consists of T7 promoter, lac operator, RBS, LL-37 coding sequence, His tag and T7 terminator, which are arranged in an order on a pET28 backbone. We aim to induce the transcription of the downstream LL-37 by adding the IPTG to initiate the expression. The protein will then be purified and antibacterial activity was tested by culturing the <i>Salmonella enterica subsp. enterica</i> (<i>ex</i> Kauffmann and Edwards) Le Minor and Popoff serovar Typhimurium with our purified LL-37 and measuring the OD<sub>600</sub> to reflect the bacteria growing condition. </p> | ||
===Expectation=== | ===Expectation=== | ||
Revision as of 14:47, 20 October 2021
LL-37 antimicrobial peptide from Humans (Homo sapiens)
LL-37 is an antimicrobial peptide (AMP) from the cathelicidin family of antimicrobial peptides, isolated from the Human (Homo sapiens). It is a 37-residue helical peptide found throughout the human body, exhibiting a broad spectrum of antimicrobial activity as well as a variety of immunomodulating effects (Dürr, Ulrich H.N., et. al 2006). It has the distinction of being the first and only cathelicidin member discovered from humans, and is extremely well studied and characterized for this reason. The peptide is produced via epithelial cells as well as leukocytes in places such as the skin, gastrointestinal tract, and the respiratory tract. This AMP is is cleaved from a larger protein, hCAP-18, in a series of modifications to yield its active form (Sorensen, O.E, et. al 2001).
Design
This part was codon optimized for E. coli using the Life Technologies GeneArt ® software program. By design, this part is Assembly Standard #23 (Silver Fusion) and contains a Methionine(atg) residue at the start of the coding region. Furthermore, this LL-37 sequence does not have any stop codons, which allows for composite BioBrick construction.
Improvement on Existing Parts (as of September 2013)
BBa_K245114 (Slovenia iGEM team 2009) - This part lacks a start codon, which would require additional design to be expressed individually. The BBa_K1162006 part improves upon this one by adding a Met(atg) residue at the beginning of the coding region. It is unclear if Slovenia's part has been codon optimized for E. coli, but our new part has been designed with this in mind by Life Technologies (see design).
BBa_K875009 (Trieste iGEM team 2012) - This part, BBa_K875009, uses the Assembly Standard #10 for both the prefix and suffix and therefore cannot be used for protein fusions, such as in cases of secretion, protein purification, or fusion proteins. This part also has a stop codon which further limits what can be added to the C-terminal end (purification tag/secretion tag). The BBa_K1162006 improves upon this part by using Assembly Standard #23 (Silver Fusion) and by removing the stop codon at the end of the coding region. Additionally, the BBa_K1162006 part was codon optimized by Life Technologies (see design).
Experience

Figure 1. The image above of protein purification fractions demonstrates that there is expression of LL-37 and WAM-1 in E. coli through GFP fluorescence (GFP is at the C-terminal end). Unfortunately, there were issues with binding to the nickel affinity column, which will be troubleshooted in future work.

Figure 2. This image is an SDS-PAGE gel of the protein products from a 10L fermentation run for the LL37-spider silk generating construct using E. coli as the host organism. In the elution fraction of the gel image below, we can see a protein band at approximately 55-60 kDa, the size of the LL37-spider silk protein. From this study we have demonstrated that antimicrobial spider silk can be produced in E. coli and scaled-up. Since we also have additional AMP-spider silk constructs (see [http://2013.igem.org/Team:Utah_State/Parts BioBricks] page for a complete list), the next step would be to manufacture more of these using a similar approach to the LL37-spider silk. In future work, optimization of LL37-spider silk production will also take place.
References
Dürr, Ulrich H.N. Sudheendra, U.S. and Ramamoorthy, Ayyalusamy, LL-37, the only human member of the cathelicidin family of antimicrobial peptides (Review). Biochimica et Biophysica Acta 1758 (2006) 1408–1425.
O.E. Sorensen, P. Follin, A.H. Johnsen, J. Calafat, G.S. Tjabringa, P.S. Hiemstra, N. Borregaard, Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3, Blood 97 (2001) 3951–3959.
Characterization: Jilin_China 2019
Group:
Jilin_China 2019Authors:
Chang YangSummary:
On the basis of BBa_K1162006, we added the termination codon and characterized it.Documention:
1. Characterization
1.1 Validation of LL-37 construction
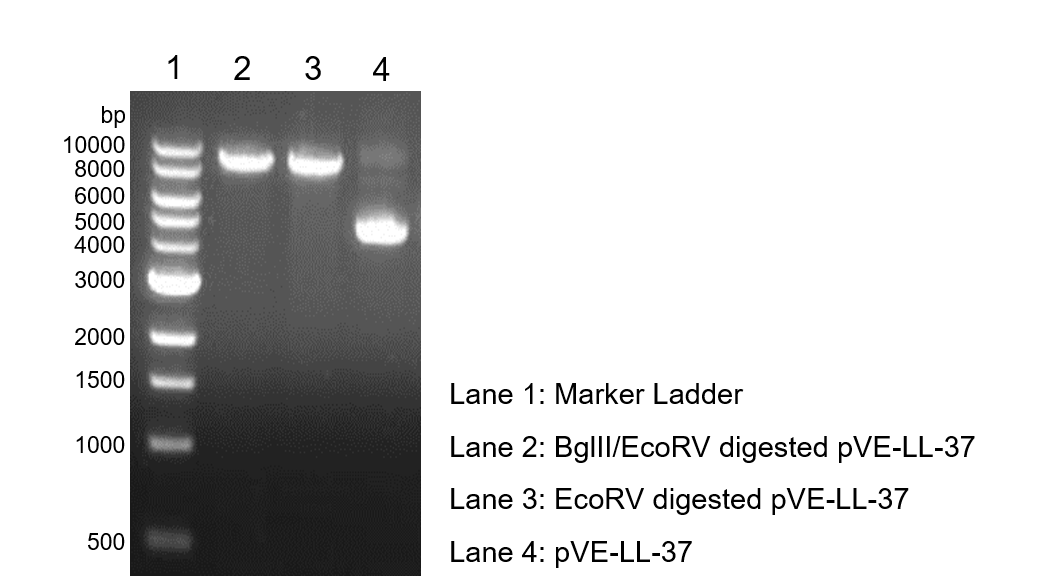
To verify the construction of pVE-LL-37 which we generated, the digestion by BglII/EcoRV was performed by a standard protocol followed by agarose gel electrophoresis (Figure 1).
Figure 1. Digestion and electrophoresis of pVE-LL-37.
1.2 Expression of pVE-LL-37
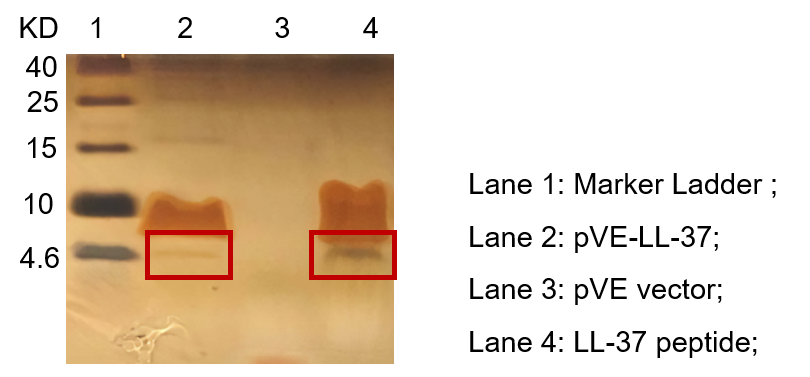
To detect the expression of pVE-LL-37 in E. coli, the constructs were transformed into BL21. Compared to the mock, there was a small size band shown in pVE-LL-37 (Figure 2).
Figure 2. Expression of pVE-LL-37. The bacteria were collected and ultrasonicated. The lysate was centrifuged and supernate was electrophoresed on the Tricine-SDS-PAGE gel, followed by Fast silver staining. The LL-37 peptide was synthesized as positive control.
1.3 Candidacidal effect of LL-37
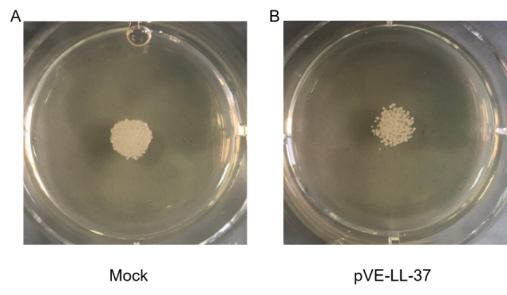
To assess the candidacidal activity of LL-37, the spot assay was performed. As shown in Figure 3, C. albicans viability after LL-37 treatment was visually compared with that of control cells (no LL-37 treatment). We found that cell viability was more sensitive to LL-37.
Figure 3. Candidacidal activity of LL-37. C. albicans were collected and suspensions were resuspended to 105 cells/mL. 10 µL C. albicans suspension and 20 µL sterile BL21-transformed bacteria were gently mixed. After a 10 min incubation, the mixture was spotted onto the SDA agar plates. Cell viability was detected after incubation at 30℃ for 18 hrs.
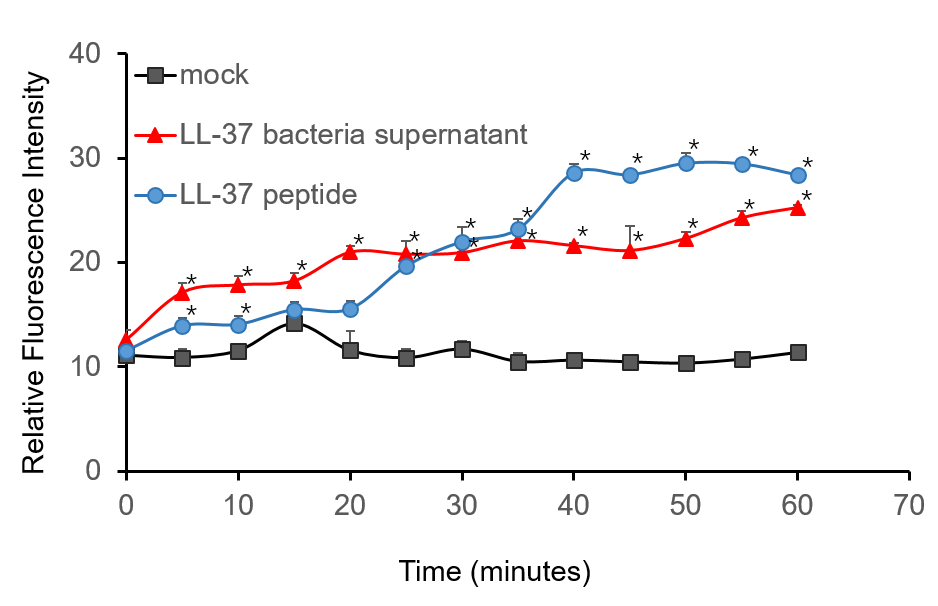
To further validate the candidacidal effect of LL-37, C. albicans suspensions were incubated with it. After incubation, LL-37 caused an immediate increase in PI fluorescence indicates that LL-37 can kill C. albicans (Figure 4).
Figure 4. Candidacidal activity of LL-37. 1.5 µM PI (propidiumiodide) was added into C. albicans suspensions (106 cells/mL, 1 mL) and serially BL21-transformed bacteria supernatant (2 mL) mixture. The whole system was incubated at 30 ℃. During the 1 h incubation, the fluorescence was measured every 5 min at the excitation wavelength λexc 544 nm and emission wavelength λem 620 nm. The experiment was performed three times in triplicate. *, P < 0.05 from control using Student’s t test.
3. Conclusion
We successfully characterized LL-37 and demonstrated its ability to kill C. albicans from multiple angles.
HZAU-China 2021
Design
In order to fit the project and guide the experiment, our research decided to depict the characterization of LL-37 protein- an antimicrobial peptide.
The plasmid we designed consists of T7 promoter, lac operator, RBS, LL-37 coding sequence, His tag and T7 terminator, which are arranged in an order on a pET28 backbone. We aim to induce the transcription of the downstream LL-37 by adding the IPTG to initiate the expression. The protein will then be purified and antibacterial activity was tested by culturing the Salmonella enterica subsp. enterica (ex Kauffmann and Edwards) Le Minor and Popoff serovar Typhimurium with our purified LL-37 and measuring the OD600 to reflect the bacteria growing condition.
Expectation
Materials and Method
Results and analysis
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]