Difference between revisions of "Part:BBa K3893030"
JeanHerdoiza (Talk | contribs) (→Part engineering and DBTL cycle) |
JeanHerdoiza (Talk | contribs) (→Learn) |
||
| Line 87: | Line 87: | ||
<b>ODE Model</b> | <b>ODE Model</b> | ||
| − | <p> | + | <p>This <a href="https://2021.igem.org/Team:Ecuador/Model#Module2">ODE Model</a> was obtained by applying the law of mass action to the system reactions and using lumped hill functions for the promoters. It has 7 states. Particularly N is the number of cells growing in the culture, and GFP is the system output. Ae is the number of molecules in the culture medium, and the remaining species are intracellular ones. |
</p><div> | </p><div> | ||
| Line 93: | Line 93: | ||
</div> | </div> | ||
<b>Computational simulation</b> | <b>Computational simulation</b> | ||
| − | <p>We learnt how our gene circuit works by comparing <b>in silico vs. in vivo results</b>. As expected, the gene circuit showed an oscillatory behaviour since cells died only when the lysis protein is activated. Therefore, | + | <p>We learnt how our gene circuit works by comparing <b>in silico vs. in vivo results</b>. As expected, the gene circuit showed an oscillatory behaviour since cells died only when the lysis protein is activated. Therefore, following the DBTL cycle we could develop a complex part (difficult to design without the DBTL cycle) and to obtain a behaviour that is compatible with the one we desired at the beggining. /p> |
<div> | <div> | ||
<img style="width: 110%" src="https://2021.igem.org/wiki/images/d/da/T--Ecuador--EngOscilatorGFP.png"> | <img style="width: 110%" src="https://2021.igem.org/wiki/images/d/da/T--Ecuador--EngOscilatorGFP.png"> | ||
Revision as of 19:59, 19 October 2021
Population control device (QS-based lysis protein oscilator)
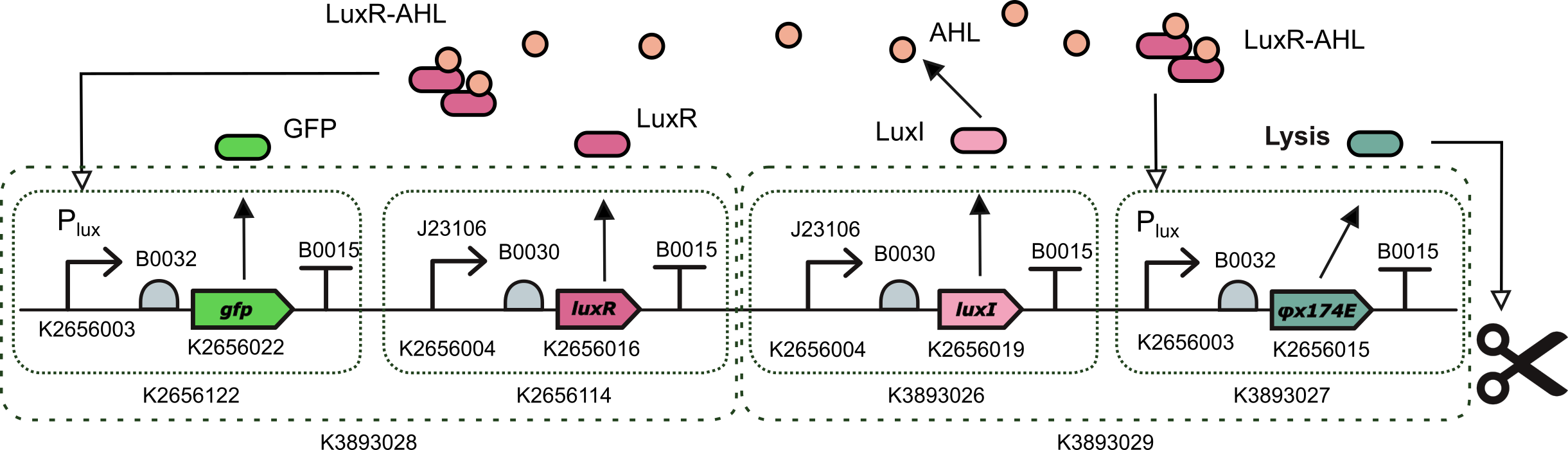
This device is an AHL induced transcriptional unit to express phiX174 lysis protein together with a TU for the constitutive expression of LuxI. It was assembled with a one-pot Level 2 Golden Gate reaction using BsmBI type IIS endonuclease.
This device is composed of the following standardized composite parts:
- BBa_K3893028: a Level 2 transcriptional unit constitutively expressing luxI gene (Golden Braid compatible)
- BBa_K3893029: a Level 2 transcriptional unit expresing phiX174 lysis protein under the control of the pux promoter (Golden Braid compatible)
Usage and Biology
Part engineering and DBTL cycle
The development of this part was performed following the steps of the Design-Build-Test-Learn (DBTL) cycle of synthetic biology. Here, we describe the different steps and the results obtained.
Design
This part is designed for delivering and releasing dsRNA molecules of the gene of interest in the context of controlled oscillations in the size of the bacterial population. However, we replaced the gene of interest by a reporter to test the effectiveness of part and to use it as a proxy to estimate the amount of lysis protein being expressed. To this end, LuxR-AHL transcription factor activates simultaneously the expression of GFP and the lysis protein PhiX174E.

Build
For this we planned a golden gate assembly with 3 levels using the Golden Braid assembly method from Valencia_UPV. First, we combined together two composite parts that were already in the Part Registry (BBa_K2656122 a Level 1 transcriptional unit expresing GFP under the control of the pux promoter and BBa_K2656114 a Level 1 transcriptional unit constitutively expressing luxR gene) to create part BBa_K3893028 .
Then we created 2 more Level 1 Transcripcional units using basic parts from the Valencia_UPV Part Collection: parts BBa_K3893026 (a Level 1 transcriptional unit constitutively expressing luxI gene) and BBa_K3893027 (a Level 1 transcriptional unit expresing phiX174 lysis protein under the control of the pLux promoter). This way, we combined these two parts and we created a new composite part BBa_K3893029.
Finally, combining these two Level 2 parts we obtained a construction with four transcriptional units implementing our initial design BBa_K3893030. For the design of part BBa_K3893026, which is a Level 1 transcriptional unit constitutively expressing luxI gene and a key component of the QS-based oscilator, we performed a smaller inned DBTL cycle including design, assembly (build), measurements (test), and model and characterization of the part (learn) using part BBa_K3893028 and contributing to characterize the pLux promoter (Part BBa_K2656003 which is the Golden Braid compatible, together with its Biobrick starndard sister part BBa_R0062) in the context of the transcriptional unit built in part BBa_K2656122.Test
We designed a temporal experiment to assess and quantify (if possible) cell lysate. We collected absorbance and fluorescence data from the gene circuit in vivo. One of the assays is shown in the Figure below.
Figures A and B depict how cells death when the lysis protein is activated by the lux promoter K2656003. The number of cells was calibrated using standardized particle units from the Engineering Committee (Measurement Committee) and the iGEM Interlab study 2018-2019.
The total GFP fluorescence expressed by the population and a single-cell is shown in Figures C and D, respectively. At the beginning of the experiment, the number of cells producing GFP is very low (OD 600=3.05e-6). But after lysis, GFP molecules are released to the medium and some cells are still producing GFP. We used MEFL/Particle (molecules of equivalent fluorescein per particle) as a standardized unit to quantify GFP expression per cell.

- All experimental measurements were taken with Biotek CytationTM 3, using a 96-well plate. The experimental conditions used in this work are in the Table below
- The cells were E. cloni® cells (Lucigen) transformed with the corresponding plasmid.
- For all experiments, we worked with an isolated colony that contains the corresponding plasmid.
- Cultures of 3mL falcon tubes with sLB medium with the corresponding antibiotic were prepared and incubated overnight (18 hours) at 37°C, 200 rpm.
- The overnight cultures were refreshed in 3 mL of sBL with the corresponding antibiotic and incubated at 37°C, 200 rpm for 4 hours.
- Culture tubes at OD600=0.05 were sit in cold water for 30 mins.
- We measured absorbance and fluorescence of four replicas of the sample starting with OD600=3.05e-6 during 14 hours.
- PROTOCOL: Time between measures 5 min, Temperature 37°C, Shaking Double orbital (Continuously), Absorbance wavelength 600 nm, Excitation wavelength 485 nm, Emission wavelength 528 nm, 96-well volume (individual) 200 μl.
Learn
We also proposed an ODE model to explain the temporal behaviour of this gene circuit. Although the ODE model only considers N=20 cells, it can qualitatively capture the dynamics of the experimental data.
ODE ModelThis ODE Model was obtained by applying the law of mass action to the system reactions and using lumped hill functions for the promoters. It has 7 states. Particularly N is the number of cells growing in the culture, and GFP is the system output. Ae is the number of molecules in the culture medium, and the remaining species are intracellular ones.

We learnt how our gene circuit works by comparing in silico vs. in vivo results. As expected, the gene circuit showed an oscillatory behaviour since cells died only when the lysis protein is activated. Therefore, following the DBTL cycle we could develop a complex part (difficult to design without the DBTL cycle) and to obtain a behaviour that is compatible with the one we desired at the beggining. /p>

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 956
Illegal NheI site found at 979
Illegal NheI site found at 1914
Illegal NheI site found at 1937 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]