Difference between revisions of "Part:BBa J23117"
(→Added by SJTU-BioX-Shanghai 2020 Team) |
|||
| Line 67: | Line 67: | ||
==Added by SJTU-BioX-Shanghai 2020 Team== | ==Added by SJTU-BioX-Shanghai 2020 Team== | ||
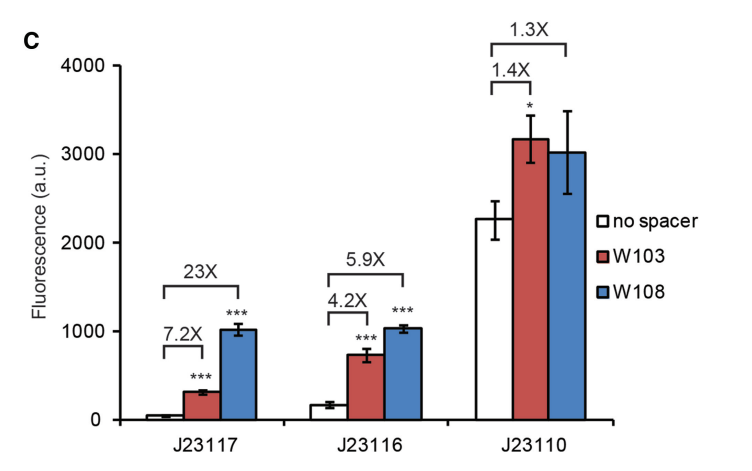
| − | + | J23117 is a weak promoter and it can lead to a quite low expression level in the absence of dCas9-ω. So, it is a good choice to be designed as a part of CRISPRa system. It will cause a significant difference in expression level of reporter after dCas9 binding. | |
[[File:Quoted_from_a_study_by_Broad_Institute.png]] | [[File:Quoted_from_a_study_by_Broad_Institute.png]] | ||
| Line 73: | Line 73: | ||
'''Figure above : Quoted from a study by Broad Institute''' | '''Figure above : Quoted from a study by Broad Institute''' | ||
| − | W103 and W108 in the figure means different ways to put the target site for dCas9-ωupstream of the promoter. Comparing the dCas9-ω activation of | + | W103 and W108 in the figure means different ways to put the target site for dCas9-ωupstream of the promoter. Comparing the dCas9-ω activation of GFP expression from the three constructs using different gRNA, J23117 performs best when induced by the dCas9-ω both in W103 and in W108. |
'''References:''' | '''References:''' | ||
[1]David Bikard,Wenyan Jiang,Poulami Samai,Ann Hochschild,Feng Zhang,Luciano A. Marraffini1.Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system[J]. Nucleic Acids Research,2013,41(15):7429-7437. | [1]David Bikard,Wenyan Jiang,Poulami Samai,Ann Hochschild,Feng Zhang,Luciano A. Marraffini1.Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system[J]. Nucleic Acids Research,2013,41(15):7429-7437. | ||
Latest revision as of 11:22, 27 October 2020
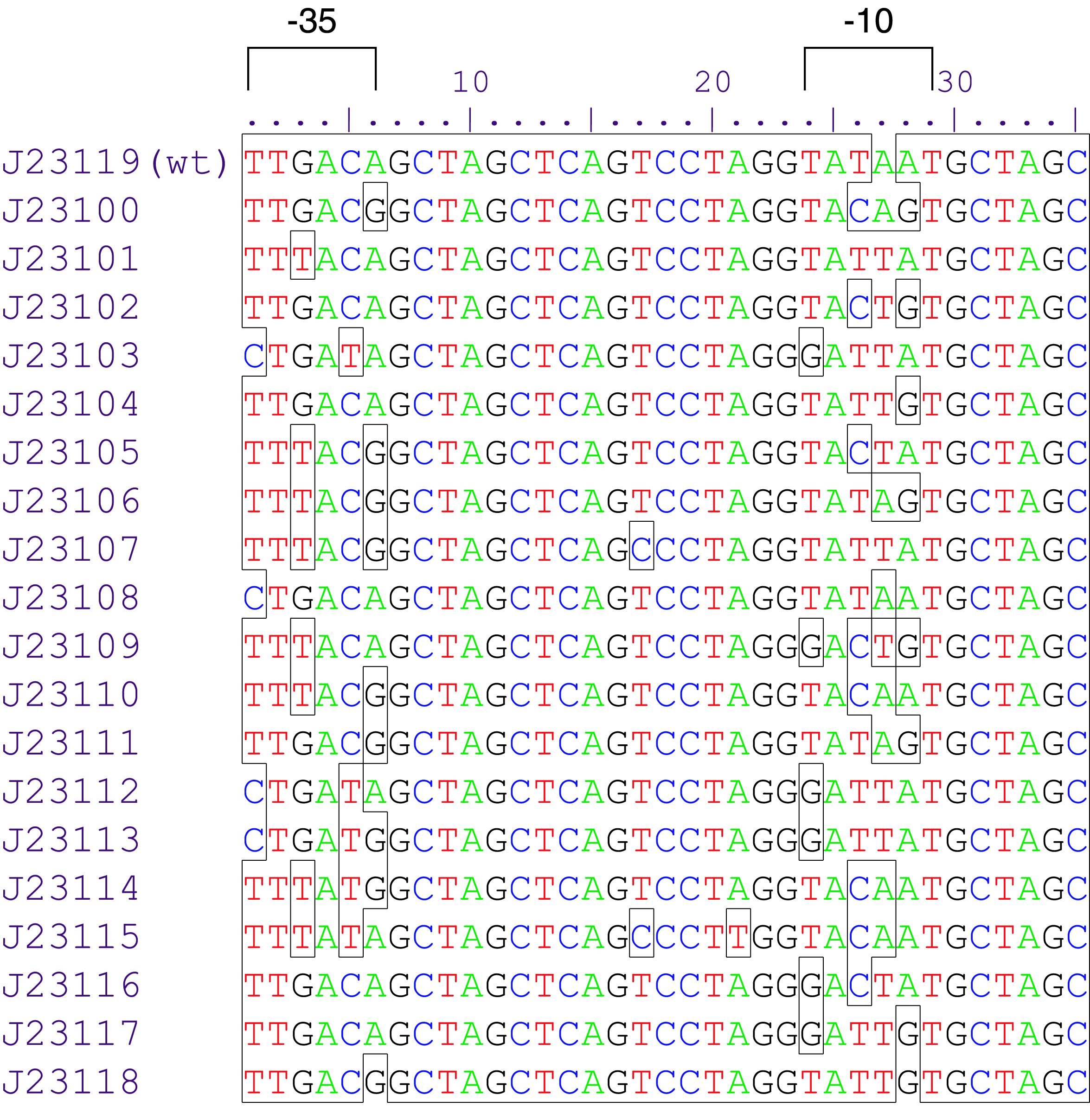
constitutive promoter family member
Variant RFP (au) J23112 1 J23103 17 J23113 21 J23109 106 J23117 162 J23114 256 J23115 387 J23116 396 J23105 623 J23110 844 J23107 908 J23106 1185 J23108 1303 J23118 1429 J23111 1487 J23101 1791 J23104 1831 J23102 2179 J23100 2547 |
Constitutive promoter family
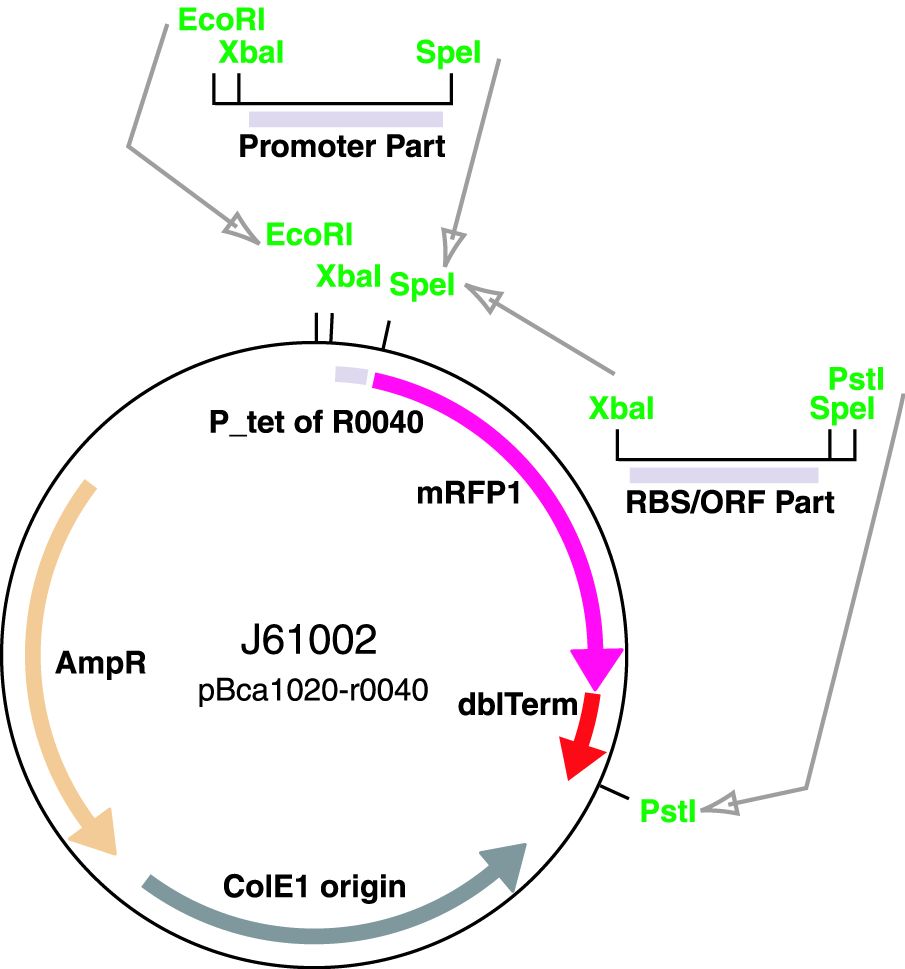
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part BBa_J61002 for details on their use.
These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. JCAraw
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
This promoter was BioBricked in 2015 competition with a PAM rich Upstream regulation sequence (URS), BBa_K1723001. This PAM rich sequence is needed for the protein dCas9 to bind to the promoter and act as a programmable transcription regulator . Another promoter, PAM rich URS J23117Alt ( BBa_K1723005 ) was created by mutating sequences of the improved J23117 promoter keeping intact -10 sequence and -35 sequence to keep a similar promoter strength.
Added by KEYSTONE_A 2020 Team
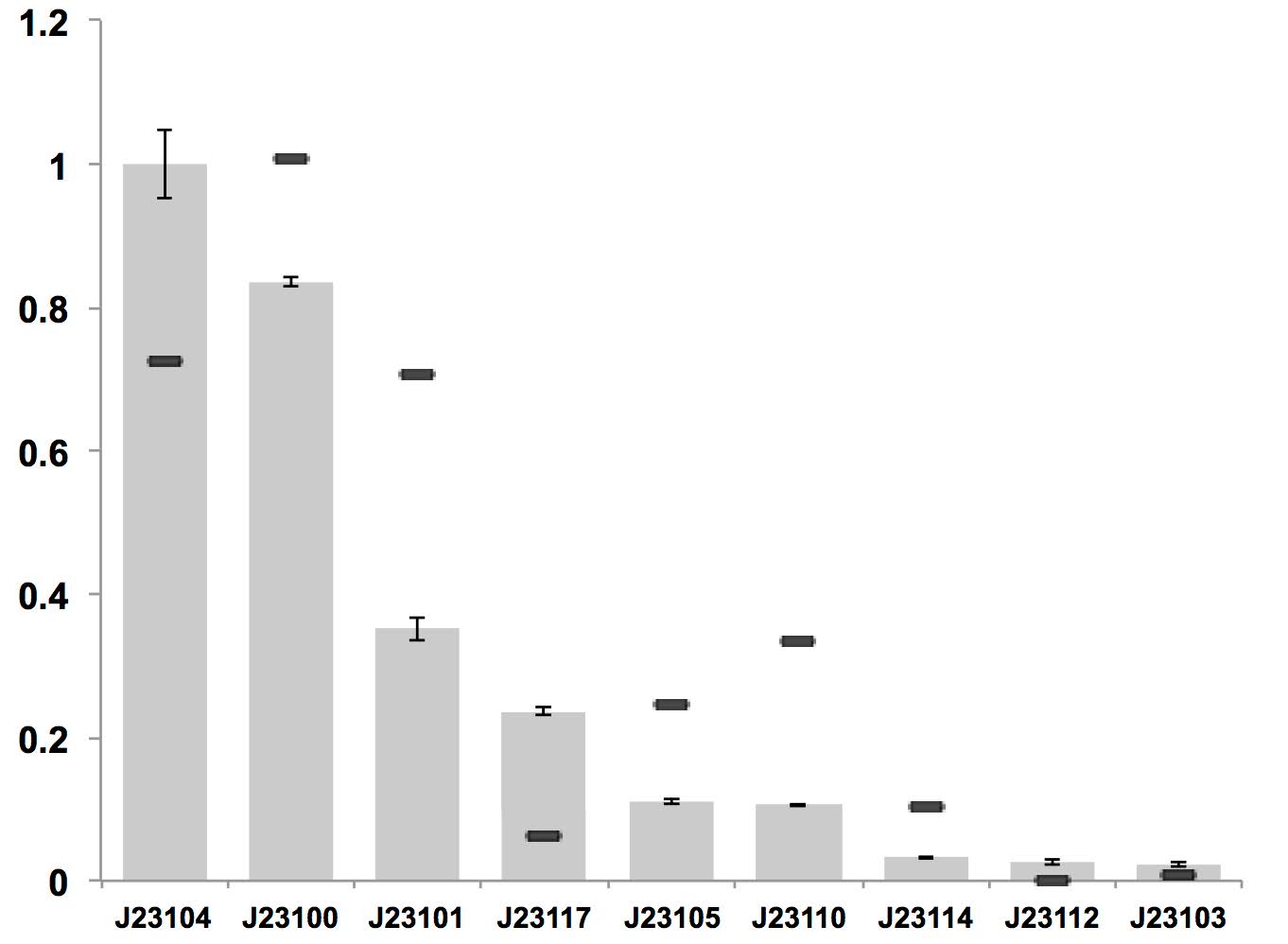
J23117 can be used as a constitutive promoter in bacterial cellulose producing strain K. rhaeticus iGEM, but it is of relatively low expression level.
Figure above: Constitutive promoter average strengths in K. rhaeticus iGEM and E. coli, normalized against J23104. Although all promoters are functional, their relative strengths differ between K. rhaeticus and E. coli. For K. rhaeticus, data is shown as grey bars, with standard deviation of N=3 biological replicates, characterized in liquid HS-medium containing cellulase, measured 3 h post-inoculation. Relative promoter strengths in E. coli are superimposed as black stripes.
References: Florea, M., Hagemann, H., Santosa, G., Abbott, J., Micklem, C. N., Spencer-Milnes, X., ... & Chughtai, H. (2016). Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proceedings of the National Academy of Sciences, 113(24), E3431-E3440.
|
•••••
University of Texas at Austin iGEM 2019 |
UT Austin iGEM 2019: Characterization of metabolic burden of the Anderson SeriesDescriptionThe 2019 UT Austin iGEM team transformed the Anderson Series promoters into our 'burden monitor' DH10B strain of E. coli, which contains a constitutive GFP cassette in the genome of the cell. GFP expression fluctuates depending on the number of ribosomes available. Using this strain, we characterized the relative burden (percent reduction in growth rate) of each Anderson Series part. Our results showed a range of growth rate reductions for each of these parts due to ribosomal reallocation from the genome of the host cell, towards the expression of RFP. Anderson Series parts with strong promoters are depicted with darker red colors and Anderson Series parts with weak promoters are depicted with lighter pink colors to show relative RFP expression. We saw a positive correlation between relative promoter strength and metabolic burden; parts with stronger promoters expressed less GFP and had a lower growth rate than parts with weaker promoters. The regression line for the graph below was constructed by measuring the burden of 5 parts that were created by the 2019 UT Austin iGEM team that each contained an Anderson Series promoter (BBa_J23104 or BBa_J23110), an RBS of varying strength, and a BFP reporter. For more information on characterization of these parts through the burden monitor, visit our team’s wiki page: [1]
Importance of Characterizing BurdenAlthough often we cannot avoid using a specific burdensome part, knowing in advance that it is burdensome, and that it has a high chance of mutating into a non-functional genetic device, can help with troubleshooting and coming up with alternatives. In the specific case of fluorescent protein-expressing devices, Fluorescence-activated cell sorting (FACS) can be used to filter out individual cells that meet a certain fluorescence threshold. This way, the cells expressing lower levels of the fluorescent protein are weeded out of the population. Added by SJTU-BioX-Shanghai 2020 TeamJ23117 is a weak promoter and it can lead to a quite low expression level in the absence of dCas9-ω. So, it is a good choice to be designed as a part of CRISPRa system. It will cause a significant difference in expression level of reporter after dCas9 binding. Figure above : Quoted from a study by Broad Institute W103 and W108 in the figure means different ways to put the target site for dCas9-ωupstream of the promoter. Comparing the dCas9-ω activation of GFP expression from the three constructs using different gRNA, J23117 performs best when induced by the dCas9-ω both in W103 and in W108. References: [1]David Bikard,Wenyan Jiang,Poulami Samai,Ann Hochschild,Feng Zhang,Luciano A. Marraffini1.Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system[J]. Nucleic Acids Research,2013,41(15):7429-7437. |