Part:BBa_K1321306
Komagataeibacter rhaeticus iGEM
Komagataeibacter rhaeticus is a obligate aerobic Gram negative cellulose-producing bacterial species. Imperial iGEM 2014 team isolated a strain of K. rhaeticus (named K. rhaeticus iGEM in the honour of the competition) from the Kombucha tea seed culture (Kombucha tea seed culture is a mixture of yeast, K. rhaeticus and other organisms, and is used to make the increasingly popular Kombucha tea). K. rhaeticus iGEM produces cellulose readily on different carbon feedstocks. Although K. rhaeticus iGEM does not produce as high amounts of cellulose as G. xylinus ATCC 53582 on glucose (see cellulose productivity on different feedstocks), its production exceeds that of ATCC 53582 on sucrose, and it is more amenable to genetic engineering and testing of genetic constructs. BBa_K1321306 is a member of the K. rhaeticus genetic engineering toolkit (parts BBa_K1321295 - BBa_K1321332).
K. rhaeticus iGEM genome sequence
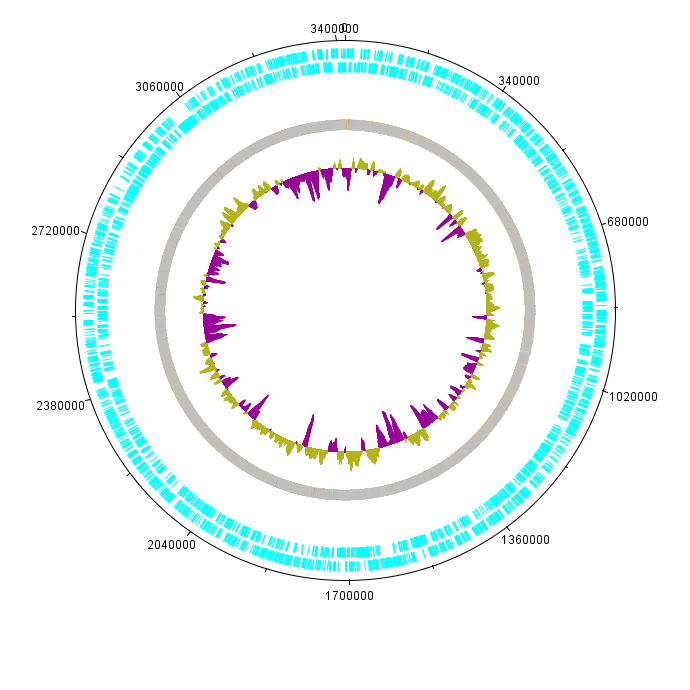
K. rhaeticus iGEM genome was sequenced as a part of Imperial iGEM 2014 project. The genome is approximately 3.4Mbp in size, with a GC content of 63.52% (see Figure 1). Autoannotation using Prokka (Seemann 2014) identified in total 3052 putative coding sequences, 59 tRNA sequences and 24 rRNA and other RNA sequences.
K. rhaeticus iGEM was sequenced using an Illumina MiSeq platform, with paired-end 250bp reads. After quality control and removal of low-quality reads, a total of approximately 0.51*10^9bp of sequenced data remained, representing approximately 150x coverage of the genome. Usually, a 50x coverage is considered sufficient for successful assembly. The full genome sequence as annotated contigs are available here File:IC14 G.xylinus igem genome.zip. This file contains the genome sequence as a fasta file and an annotated gff file (Prokka output, which requires Artemis genome browser for viewing). Note, that these files contain contigs, which are not yet ordered into a scaffold. This is a precaution to avoid misrepresentation of the large-scale organization of the genome, as the genomes of the closest relatives have proven to be inappropriate for scaffolding. This does not however, affect the quality of the sequences themselves. Note that although the iGEM strain was initially thought to be a strain of Gluconacetobacter xylinus, a later analysis using 16s rRNA places it nearest to Komagataeibacter rhaeticus, meaning that iGEM likely belongs to this species.
Culturing K. rhaeticus iGEM
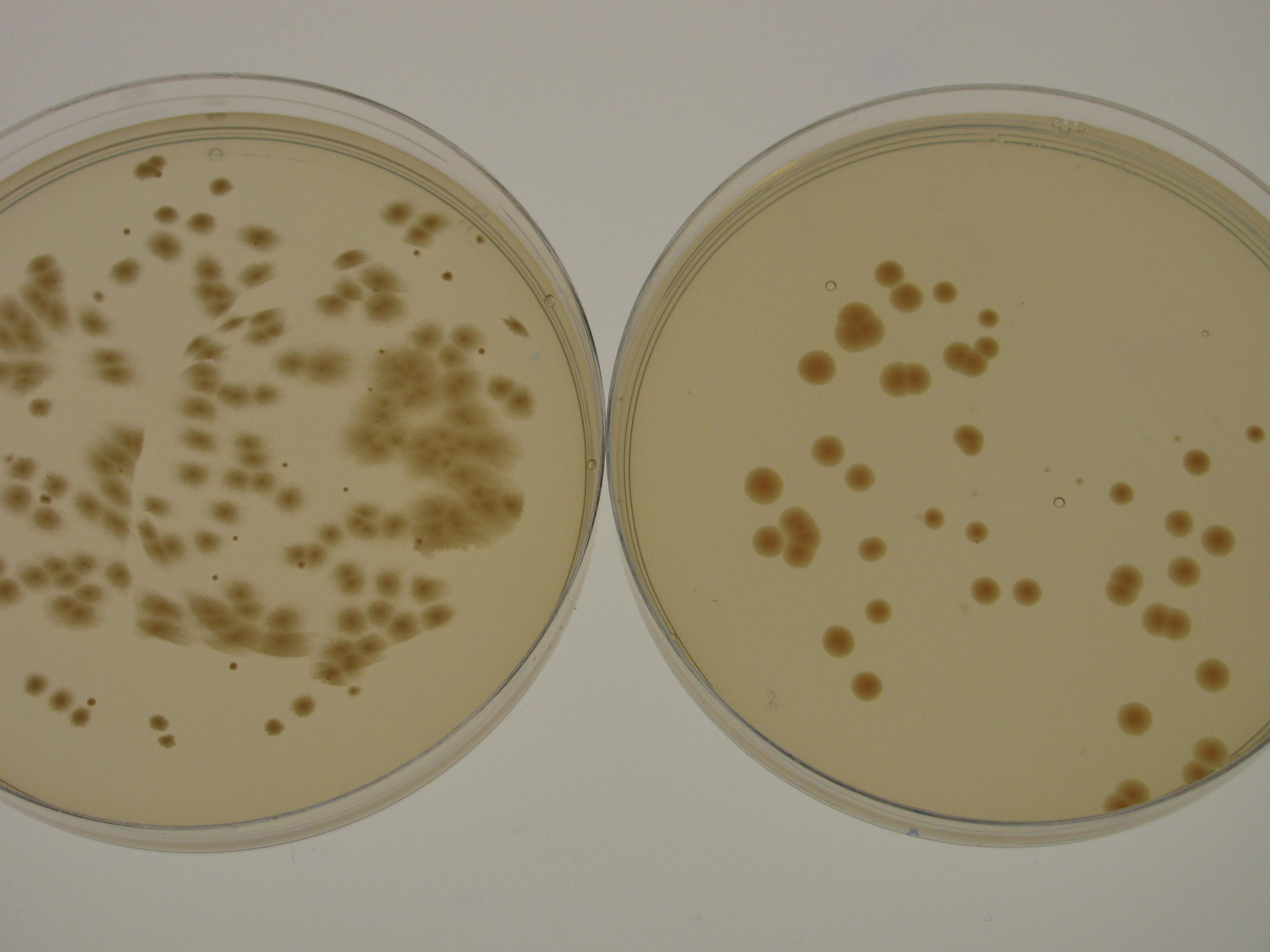
K. rhaeticus iGEM growth rate varies depending on the culturing conditions. Growth rate is highest when K. rhaeticus is cultured at 30degC, 180rpm shaking in a well-aerated setting. However, in shaking conditions, the prevalence of cellulose non-producing mutants (cel- mutants) increases, which causes a decrease in the total yield of produced cellulose. For this reason, when high cellulose production is required, K. rhaeticus is often grown in static conditions, at 30degC, in a well-aerated setting. In shaking cultures, iGEM strain reaches stationary phase in 48-72 hours, whereas in static cultures, depending on the volume of the medium, it can take up to 15 days. When grown on HS-agar plates at 30degC, cellulose-producing colonies become visible within 48-72 hours, cel- mutant colonies between 24 and 48 hours. Cellulose producing colonies can be distinguished from cel- mutants by colony morphology: cel+ have a rough colony morphology, whereas cel- mutants have a smooth colony morphology (see Figure 2).
The most commonly used medium for "K. rhaeticus" culturing is Hestrin-Schramm (HS) medium. HS medium contains the following components (for a detailed overview for K. rhaeticus culturing, see G. xylinus culturing on [http://2014.igem.org/Team:Imperial/Protocols Imperial 2014 wiki]:
2% (w/v) glucose
0.5% (w/v) yeast extract
0.5% (w/v) peptone
0.27% (w/v) Na2HPO4
0.15% (w/v) citric acid.
Cellulose productivity of K. rhaeticus iGEM
K. rhaeticus iGEM cellulose productivity depends strongly on the carbon feedstocks used. In the lab, K. rhaeticus is commonly grown on HS-glucose. In HS-glucose media grown for 7 days at 30degC standing using 50ml HS medium in 250ml conical flasks (sealed with foam buns), cellulose productivity is approximately half of that of ATCC 53582 strain (see Figure 3 for comparison of ATCC53582 and iGEM strains and experimental details). However, it can readily use other carbon feedstocks, including sucrose and glycerol (not shown).However, iGEM growth is not optimal in HS-glucose, because in HS-sucrose media the cellulose productivity is increased. Furthermore, in HS-sucrose medium, cellulose productivity is higher than that of ATCC 53582, which is of industrial significance due to the high availability of sucrose.
Natural antibiotics resistance of K. rhaeticus iGEM
Using optimal antibiotics concentrations with K. rhaeticus iGEM is critical for selection of transformed cells while not overly inhibiting cell growth. K. rhaeticus iGEM is naturally resistant to kanamycin, ampicillin and chloramphenicol concentrations normally used for E.coli (50µg/ml, 100µg/ml and 35µg/ml respectively; see Figure 4 for an experiment of iGEM natural antibiotic concentrations, experimental details, and comparison of ATCC and iGEM strains). Imperial iGEM 2014 team found that iGEM strain forms antibiotic-resistant colonies up to 6x antibiotic concentrations on HS-agarose (6x of those commonly used for E.coli), thus the recommended amounts of antibiotics for HS-plates after transformations are 350µg/ml kanamycin, 700µg/ml ampicillin and 245µg/ml chloramphenicol.
Plasmid backbones for K. rhaeticus iGEM
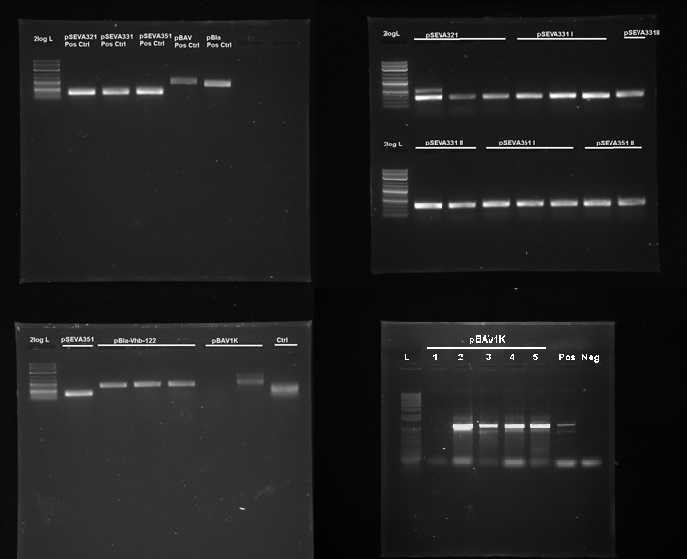
We tested 9 different plasmid backbones previously not known to replicate in Acetobacteraceae - pSEVA311, pSEVA321, pSEVA331, pSEVA341, pSEVA351, pBla-Vhb-122 (from Chien et al. 2006), pBAV1K-T5-sfgfp, pSB1C3 and pBca1020 (part number BBa_J61002) in order to create a set of plasmid backbones with different copy numbers for K. rhaeticus genetic engineering. We used pBla-Vhb-122 as the positive control, as it has been previously shown to replicate in G. xylinus (see Chien et al. 2006). In order to test for replication, we prepared electrocompetent cells (see Preparing electrocompetent cells) and transformed using 33µl, 50µl and 100µl of electrocompetent cells and 50µg, 200µg and 500µg of DNA (see Transformation of G. xylinus using electroporation). Transformants were plated out on 4 different antibiotic concentrations for each transformation: 0.5x, 1x, 2x and 6x concentrations (this refers to concentrations of antibiotics commonly used for E. coli, which are 50µg/ml kanamycin, 100µg/ml ampicillin and 35µg/ml chloramphenicol). Transformed colonies could readily be achieved. Colonies were then inoculated into HS-cellulase medium and grown for 3 days shaking at 180 RPM with appropriate antibiotics, after which plasmid DNA was miniprepped using the QiaPrep Spin Miniprep kit, according to manufacturer's instructions. Miniprepped DNA was then used as a template for PCR to determine plasmid-containing colonies. Primers used for PCR had been previously verified with pure plasmid DNA. DNA sequence of successfully amplified plasmids were confirmed by Sanger sequencing.
We discovered that 5 out of the 9 tested plasmids - pSEVA321, pSEVA331, pSEVA351, pBAV1K and pBla-Vhb-122 (as the positive control)- were capable of replication in both K. rhaeticus iGEM (see Figure 5). No positive results were obtained for other plasmids. As we tested multiple antibiotic and DNA concentrations, this indicates that the rest of the plasmids can not replicate in K. rhaeticus due to incompatible origin of replications. We converted pSEVA321 and pSEVA331 into a biobrick-compatible format by inserting the biobrick prefix and suffix in place of the original multiple cloning site using PCR with mutagenic primers and are currently in the process of converting pSEVA351 and pBla-Vhb-122 (converting them into biobrick format has required more time, as they contain several forbidden restriction enzyme sites requiring multiple steps of mutagenesis). pBAV1K was already engineered to be biobrick compatible by authors. We have also verified replication of all of these plasmids in E.coli. Thus, we have determined 5 new plasmid backbones that can act as shuttle vectors for K. rhaeticus genetic engineering:
pSEVA321Bb - part BBa_K1321301 pSEVA331Bb - part BBa_K1321300 pSEVA351 - part BBa_K1321307 pBla-Vhb-122 - part BBa_K1321308 pBAV1K - part BBa_K1321309
NOTE: Because the registry's standard plasmid backbone pSB1C3 is not capable of replication in Acetobacteracae species, the K. rhaeticus genetic engineering toolkit is housed mainly in pSEVA331-Bb. pSEVA331-Bb is a non-standard backbone, which therefore can't be quality controlled by and maintained in the Registry. However, in order to make the K. rhaeticus toolkit available for the synthetic biology community, Imperial iGEM 2014 team has made it freely available upon request, with quality control provided (see Experience). To request, please contact Imperial iGEM 2014 team.
References:
Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England). [Online] 30 (14), 2068–2069. Available from: doi:10.1093/bioinformatics/btu153 [Accessed: 10 July 2014].
Added by KEYSTONE_A 2020 Team
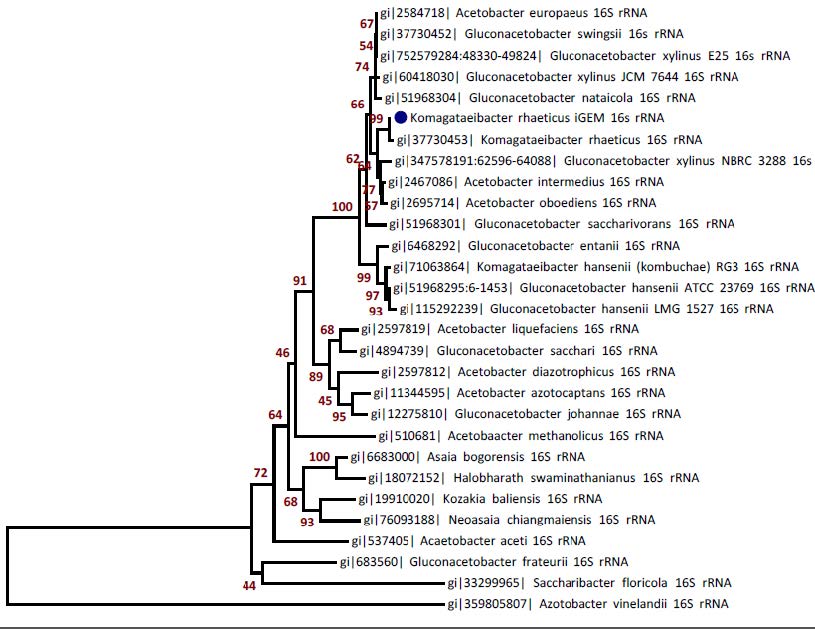
The genome sequence revealed several interesting aspects about the biology of K. rhaeticus iGEM. First, a 16S rRNA phylogeny suggests the iGEM strain to be a previously unidentified strain of K. rhaeticus, rather than G. xylinus, which is normally thought to be associated with Kombucha tea.
Figure above: 16s rRNA phylogeny of K. rhaeticus iGEM with 29 Acetobacteraceae species. Blue dot indicates the iGEM strain.
Although the strain was initially described as Gluconacetobacter xylinus by the providers of the Kombucha culture, the phylogeny instead places the iGEM strain closest to Komagataeibacter rhaeticus. This is supported by the high percent identity (99.93%) between the iGEM and K. rhaeticus 16S rRNA, which together indicate that iGEM is a new strain of K. rhaeticus. Nucleotide sequences were aligned and percent identity calculated using MUSCLE and the tree was generated using the Neighbour-Joining method. The tree is drawn to scale, with Bootstrap values from 1000 replicates shown next to the branches. Nucleotide sequences and all positions containing gaps and missing data were eliminated. Analyses were conducted in MEGA6 package.
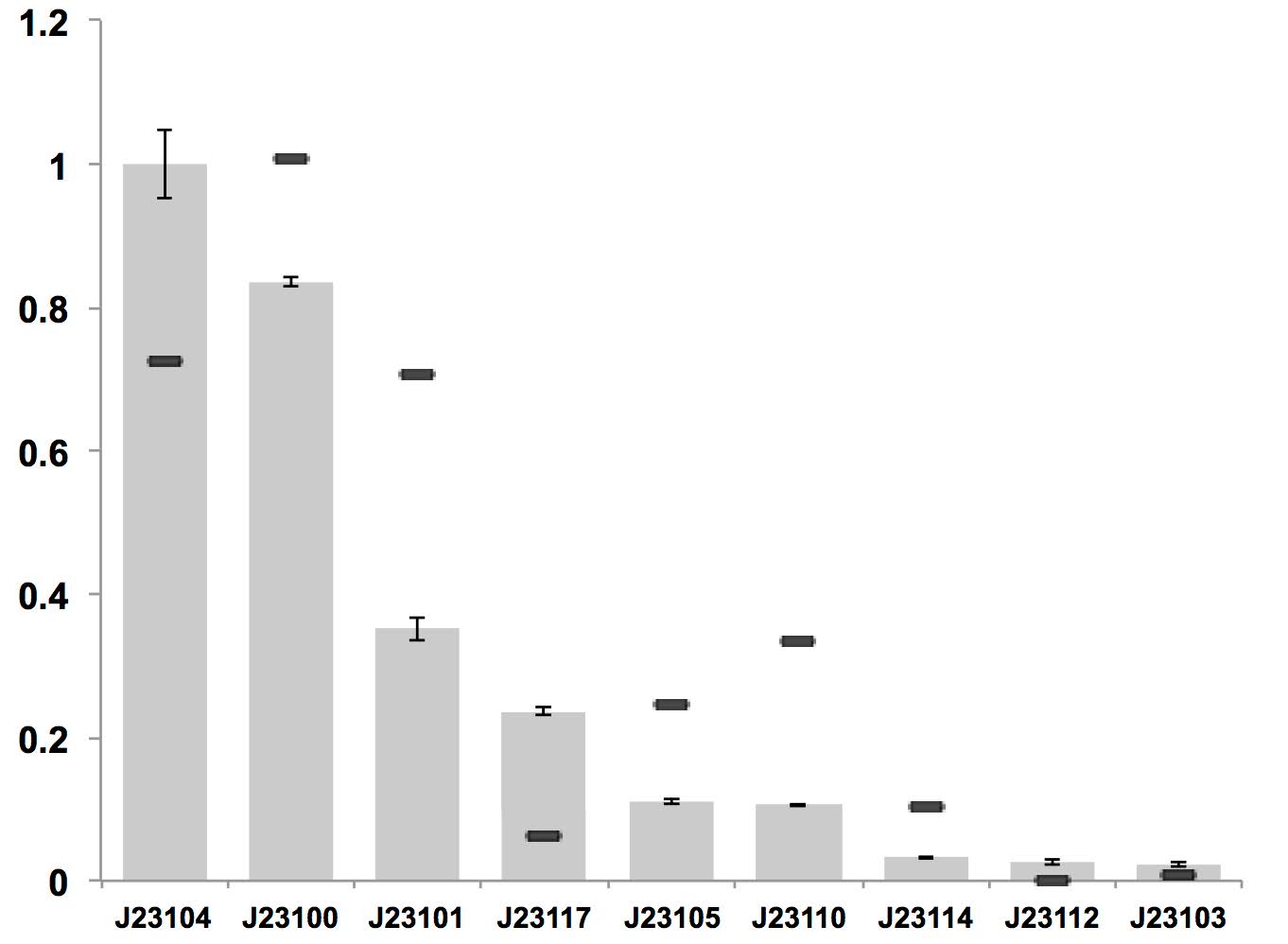
A constitutive promoter library has been established for strain K. rhaeticus iGEM, which is based on constitutive promoter family J23119 from E. coli.
Figure above: Constitutive promoter average strengths in K. rhaeticus iGEM and E. coli, normalized against J23104.
Although all promoters are functional, their relative strengths differ between K. rhaeticus and E. coli. For K. rhaeticus, data is shown as grey bars, with standard deviation of N=3 biological replicates, characterized in liquid HS-medium containing cellulase, measured 3 h post-inoculation. Relative promoter strengths in E. coli are superimposed as black stripes, and taken from existing online data (23) where promoter strengths were also characterized using mRFP1.
References:
Florea, M., Hagemann, H., Santosa, G., Abbott, J., Micklem, C. N., Spencer-Milnes, X., ... & Chughtai, H. (2016). Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proceedings of the National Academy of Sciences, 113(24), E3431-E3440.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |