Difference between revisions of "Part:BBa K1031912"
| Line 46: | Line 46: | ||
Characterized by BHSF_ND 2019 | Characterized by BHSF_ND 2019 | ||
| − | |||
The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). | The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). | ||
| − | |||
| − | As the result shows, | + | As the result shows, the system exhibit relative high level of fluorescence compared to our backbone design( BN006/Contains BBa_K3202029) which suggest the inducible transcription system function properly. |
| − | + | ||
| − | + | ||
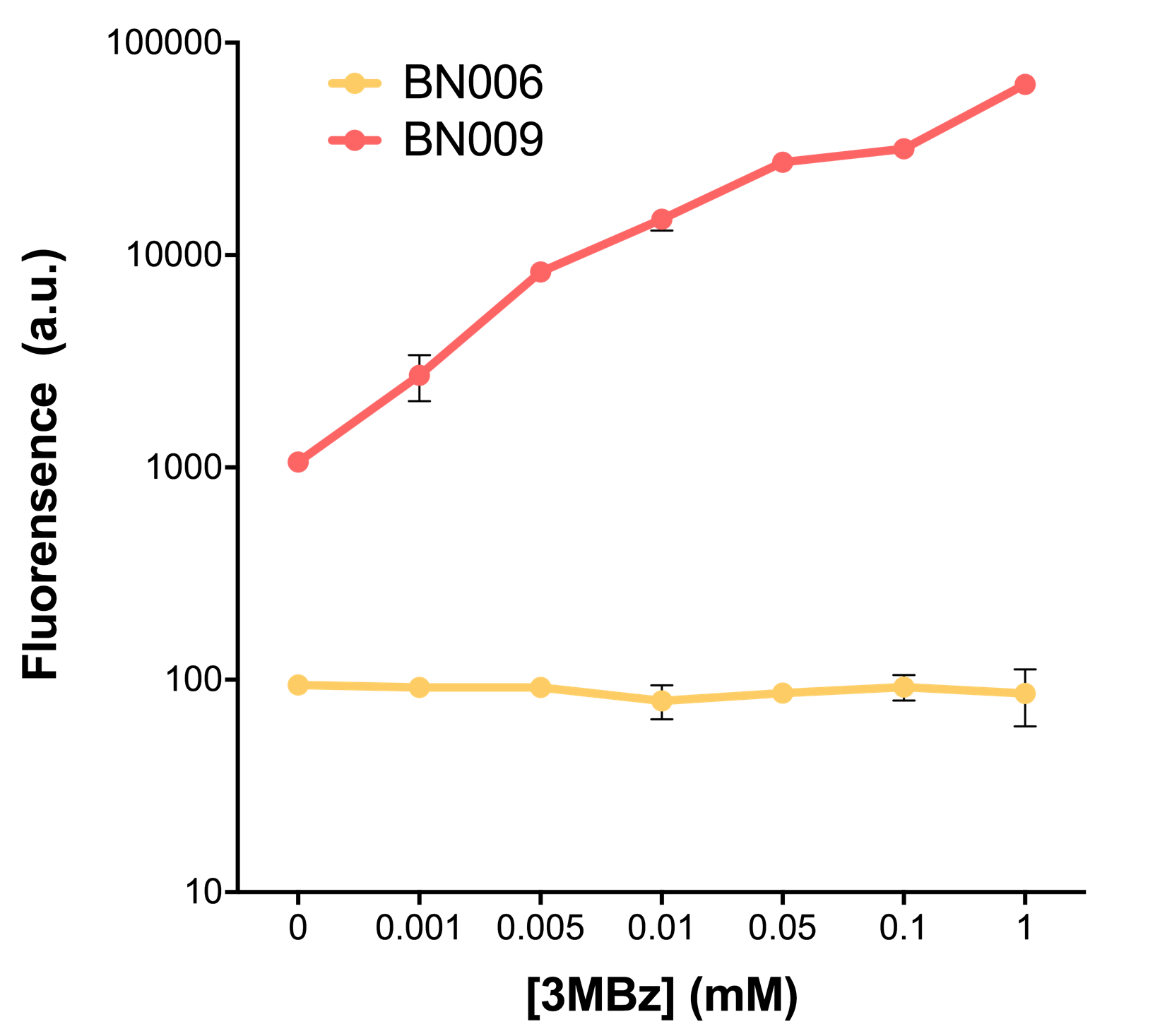
[[File:T--BHSF_ND--Leakage experiment 2.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure 2: Xyls(BN009/Contains BBa_K3202031) induction curve at different concentrations after 4 hours compared to backbone(BN006/Contains BBa_K3202029)]] | [[File:T--BHSF_ND--Leakage experiment 2.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure 2: Xyls(BN009/Contains BBa_K3202031) induction curve at different concentrations after 4 hours compared to backbone(BN006/Contains BBa_K3202029)]] | ||
| − | |||
| − | |||
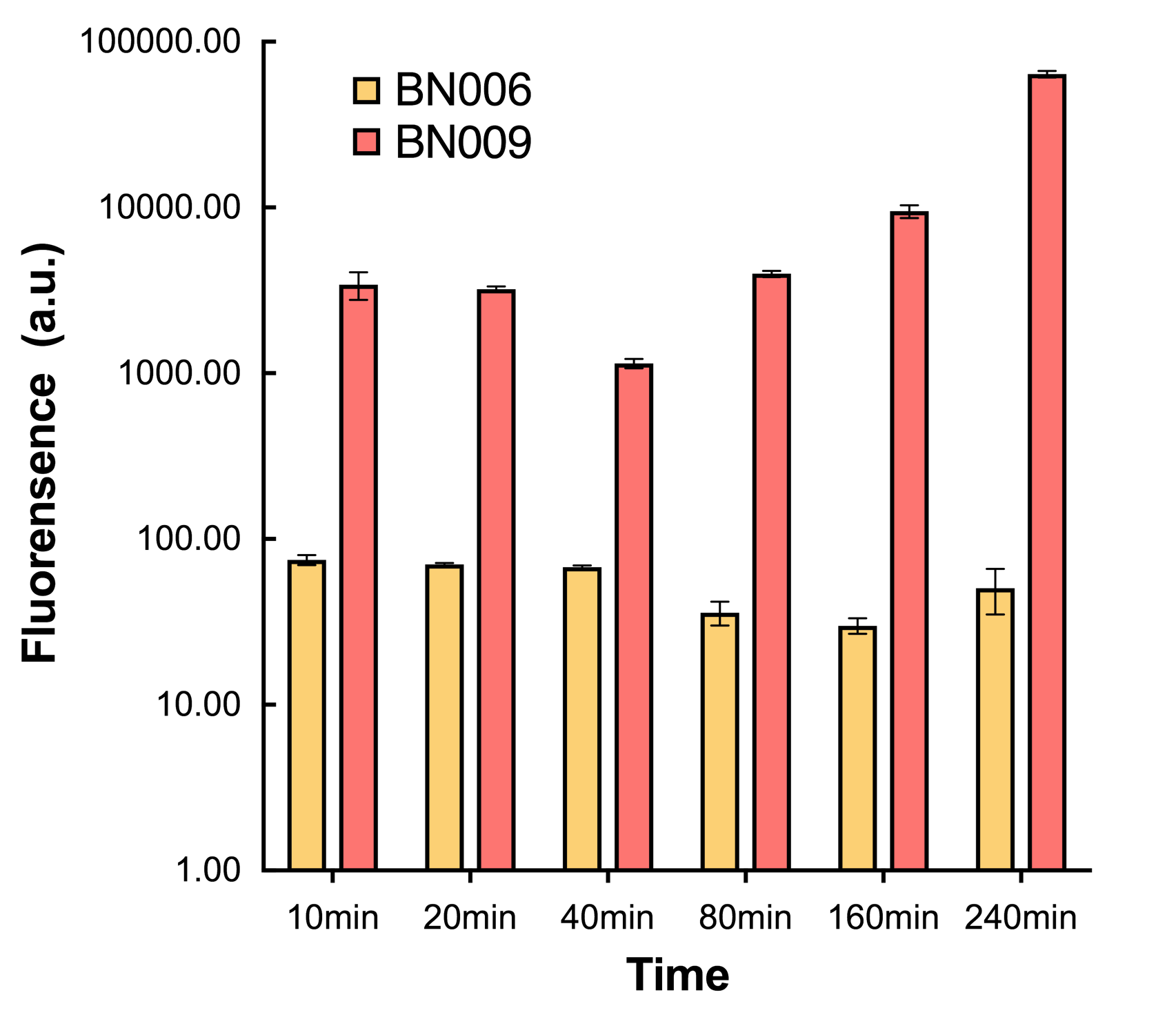
[[File:T--BHSF_ND--Leakage experiment 4.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure4: Time induction curve for Xyls(BN009/Contains BBa_K3202031) at the concentration of 1mM compared to backbone (BN006/Contains BBa_K3202029)]] | [[File:T--BHSF_ND--Leakage experiment 4.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure4: Time induction curve for Xyls(BN009/Contains BBa_K3202031) at the concentration of 1mM compared to backbone (BN006/Contains BBa_K3202029)]] | ||
| Line 141: | Line 135: | ||
In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication. | In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication. | ||
| − | + | The inducers and its respective promoters (Xyls & Pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducer (3MBz) is present, promoter (Pm) is initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. | |
| − | When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP) | + | When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP). The result suggested that the system have noticeable leakage. |
| − | When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of | + | When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of Xyls have increased exponentially. This is another sign that the system function properly. |
| − | We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone | + | We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry. |
Revision as of 23:10, 21 October 2019
J23105-XylS-Terminator
For detailed information concerning XylS, please visit 2013 Peking iGEM Biosensor XylS

Characterization
In order to fine-tune the performance of XylS biosensor, we constructed a library of Pc constitutive promoters at different intensity. BBa_K1031912 is composed of three elements, the constitutive Pc promoter J23105, coding sequence of XylS and terminator B0015. (Fig 1)

Fig 1 Construction of biosensor circuit Pm/J23105-XylS. Orange arrowheads represent promoters. RBSs are shown as green ovals and squares in dark red stand for B0015 terminator.
[http://2019.igem.org/Team:BHSF_ND BHSF_ND 2019] Characterised the efficiency on the function of translating fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration.Also, we find out that at a certain time node that the system will suddenly increase its productivity. At last, the result suggested that both system have noticeable leakage and Xyls has much higher leakage. Check results on Experience.
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 93
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 815
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 278
Data shown
Induction ratio of XylS biosensor library from ON/OFF test that utilizes different Pc promoters to control the expression of XylS. The dashed box refers to data for Pm/J23105-XylS biosensor circuit (Fig 2)

Fig 2 Horizontal axis stands for different XylS biosensor with Pc promoters of different strength. The expression strength of these constitutive promoters, J23113, J23109, J23114, J23105, J23106 is 21, 106, 256, 623, and 1185, respectively, according to the Parts' registry. Four kinds of aromatics, namely BzO, 2-MxeBzO, 3-MeBzO and 4-MeBzO, shown with different color intensities, were tested following Test Protocol 1. Vertical axis represents the ON/OFF induction ratio. The dashed box refers to performance for Pm/J23105-XylS biosensor circuit.
The performance of two inducible promoters(AraC-pBAD compared to Xyls-Pm) in terms of time and concentration variation
Characterized by BHSF_ND 2019
The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design).
As the result shows, the system exhibit relative high level of fluorescence compared to our backbone design( BN006/Contains BBa_K3202029) which suggest the inducible transcription system function properly.
Results
In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication.
The inducers and its respective promoters (Xyls & Pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducer (3MBz) is present, promoter (Pm) is initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP). The result suggested that the system have noticeable leakage.
When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of Xyls have increased exponentially. This is another sign that the system function properly.
We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry.