Difference between revisions of "Part:BBa K3110007"
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3110007 short</partinfo> | <partinfo>BBa_K3110007 short</partinfo> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
| Line 17: | Line 12: | ||
<partinfo>BBa_K3110007 parameters</partinfo> | <partinfo>BBa_K3110007 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | <h1>Usage and Biology</h1> | ||
| + | |||
| + | This construct has L-Lactate Dehydrogenase (lldD) under the control of a medium promoter and a strong RBS. | ||
| + | lldD cleaves L-lactate to pyruvate. Our intention of using this part is to have control over the detection threshold of L-Lactate by the lldPRD regulatory region. We have also designed other parts which produce various higher and lower concentrations of lldD by varying the strength of the promoter and RBS controlling its production. | ||
| + | |||
<h1>Characterization of the Part</h1> | <h1>Characterization of the Part</h1> | ||
| + | |||
| + | |||
| + | First we chose only lldRO1-J23117-lldRO2 Strong RBS sfGFP to distinguish between cancerous and non-cancerous micro-environments. | ||
| + | |||
| + | [[File:T--IISER Tiruapti--strong.png|thumb|center|700px|Figure 1. Relative Expression of sfGFP under the control of lldRO1-J23117-lldRO2 ([[Part:BBa_K1847008]]) and Constitutive Promoter with Strong RBS.]] | ||
| + | |||
| + | |||
| + | From the above graph it's clear that the construct lldRO1-J23117-lldRO2 Strong RBS sfGFP ([[Part:BBa_K3110040]]) gives a sigmoidal pattern for detection of lactate with a threshold of around 0.45 mM. However, this is a bad threshold since the construct might fail to distinguish Cancerous vs Non-cancerous microenvironments, both of which have lactate concentration than 0.45mM. Hence we have intended to use this construct in combination with our library of four gene constructs: lldD, lldP+lldD, lldR+lldD and lldP+lldR+lldD under promoter and RBS of different strengths. | ||
| + | |||
| + | We hypothesized that, if we can increase the amount of Lactate Dehydrogenase inside the cell, it will cleave some amount of Lactate. So, the surge in sfGFP expression should be seen in a higher concentration of lactate induction. | ||
| + | |||
| + | <h3>Genetic Design for the Characterization</h3> | ||
| + | |||
| + | So, we designed the following gene construct to produce more LldD (L-Lactate dehydrogenase) in a different plasmid (pSB3K3): | ||
| + | <bold>Medium Promoter-Strong RBS-lldD-Terminator</bold> | ||
| + | We used medium promoter and cloned into a low copy number plasmid so that the amount of lldD inside the cell shouldn’t be too high to cleave all the lactate inside the cell and there is no lactate left inside the cell to induce O1PO2. Also, pSB3K3 and pSB1C3 are compatible plasmids and hence can be co-transformed. | ||
| + | Then we co-transformed the 3 constructs in BL21(DE3) in 2 different combinations: | ||
| + | <ul> | ||
| + | <li>Medium P-Strong RBS-lldD (in pSB3K3) + O1PO2-Strong RBS-sfGFP (in pSB1C3) | ||
| + | <li>Medium P-Strong RBS-lldD (in pSB3K3) + P-Strong RBS-sfGFP (in pSB1C3) | ||
| + | </ul> | ||
| + | |||
| + | <h3>Results</h3> | ||
[[File:T--IISER Tiruapti--lldD.png|thumb|center|700px|Figure 1. Relative Expression of sfGFP under the control of lldRO1-J23117-lldRO2([[Part:BBa_K1847008]])-strong RBS and Constitutive Promoter-strong RBS along with Medium Promoter-Strong RBS-lldD.]] | [[File:T--IISER Tiruapti--lldD.png|thumb|center|700px|Figure 1. Relative Expression of sfGFP under the control of lldRO1-J23117-lldRO2([[Part:BBa_K1847008]])-strong RBS and Constitutive Promoter-strong RBS along with Medium Promoter-Strong RBS-lldD.]] | ||
| + | |||
| + | From the above | ||
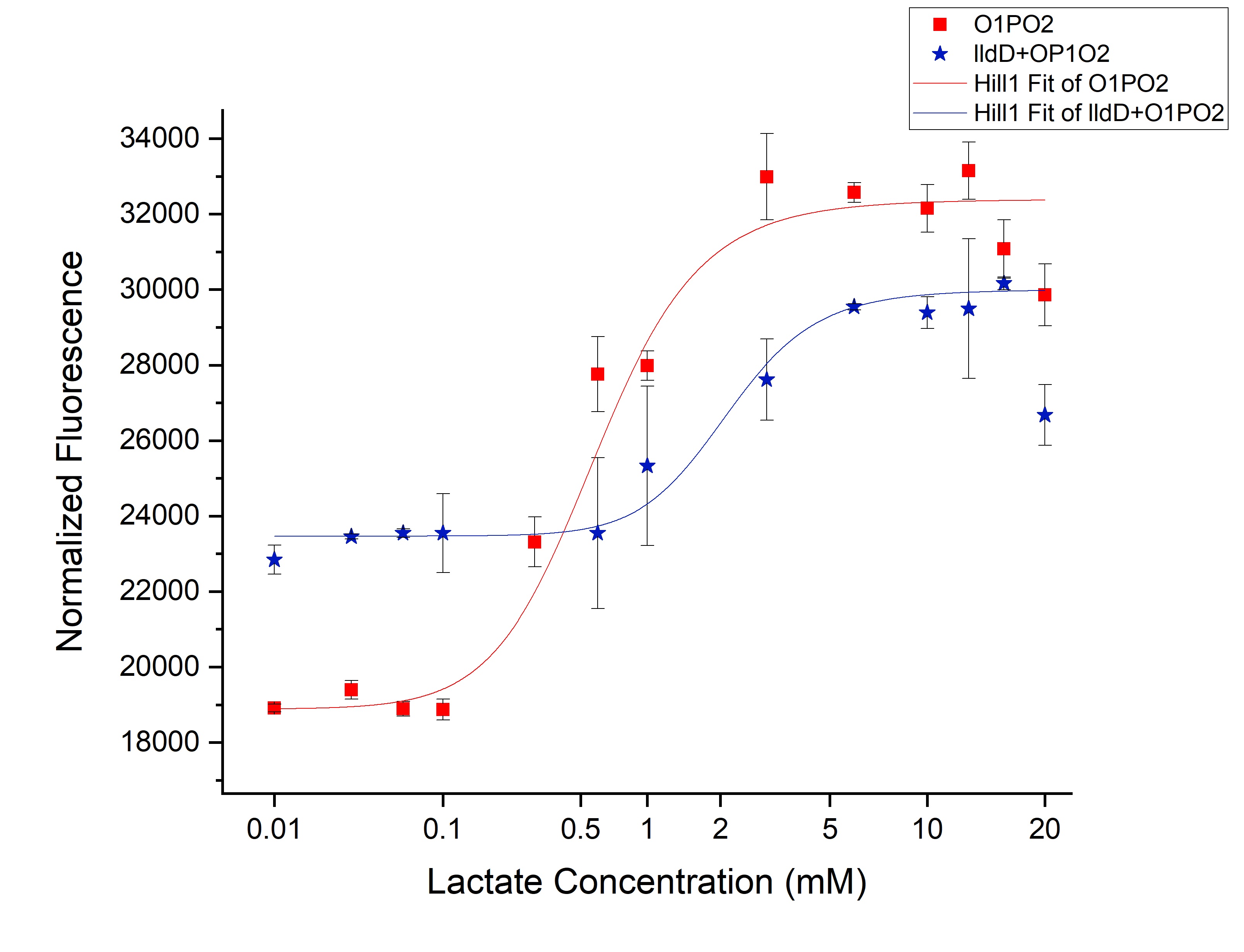
<h3>Comparison between only O1PO2 and O1PO2 along with lldD</h3> | <h3>Comparison between only O1PO2 and O1PO2 along with lldD</h3> | ||
[[File:T--IISER Tirupati--compare.png|thumb|center|700px|Figure 1. Relative Expression of sfGFP under the control of only lldRO1-J23117-lldRO2([[Part:BBa_K1847008]])-strong RBS and lldRO1-J23117-lldRO2-strong RBS along with Medium Promoter-Strong RBS-lldD ([[BBa_K3110007]]).]] | [[File:T--IISER Tirupati--compare.png|thumb|center|700px|Figure 1. Relative Expression of sfGFP under the control of only lldRO1-J23117-lldRO2([[Part:BBa_K1847008]])-strong RBS and lldRO1-J23117-lldRO2-strong RBS along with Medium Promoter-Strong RBS-lldD ([[BBa_K3110007]]).]] | ||
Revision as of 11:29, 21 October 2019
Medium Promoter Strong RBS lldD
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1202
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 639
Usage and Biology
This construct has L-Lactate Dehydrogenase (lldD) under the control of a medium promoter and a strong RBS. lldD cleaves L-lactate to pyruvate. Our intention of using this part is to have control over the detection threshold of L-Lactate by the lldPRD regulatory region. We have also designed other parts which produce various higher and lower concentrations of lldD by varying the strength of the promoter and RBS controlling its production.
Characterization of the Part
First we chose only lldRO1-J23117-lldRO2 Strong RBS sfGFP to distinguish between cancerous and non-cancerous micro-environments.

From the above graph it's clear that the construct lldRO1-J23117-lldRO2 Strong RBS sfGFP (Part:BBa_K3110040) gives a sigmoidal pattern for detection of lactate with a threshold of around 0.45 mM. However, this is a bad threshold since the construct might fail to distinguish Cancerous vs Non-cancerous microenvironments, both of which have lactate concentration than 0.45mM. Hence we have intended to use this construct in combination with our library of four gene constructs: lldD, lldP+lldD, lldR+lldD and lldP+lldR+lldD under promoter and RBS of different strengths.
We hypothesized that, if we can increase the amount of Lactate Dehydrogenase inside the cell, it will cleave some amount of Lactate. So, the surge in sfGFP expression should be seen in a higher concentration of lactate induction.
Genetic Design for the Characterization
So, we designed the following gene construct to produce more LldD (L-Lactate dehydrogenase) in a different plasmid (pSB3K3): <bold>Medium Promoter-Strong RBS-lldD-Terminator</bold> We used medium promoter and cloned into a low copy number plasmid so that the amount of lldD inside the cell shouldn’t be too high to cleave all the lactate inside the cell and there is no lactate left inside the cell to induce O1PO2. Also, pSB3K3 and pSB1C3 are compatible plasmids and hence can be co-transformed. Then we co-transformed the 3 constructs in BL21(DE3) in 2 different combinations:
- Medium P-Strong RBS-lldD (in pSB3K3) + O1PO2-Strong RBS-sfGFP (in pSB1C3)
- Medium P-Strong RBS-lldD (in pSB3K3) + P-Strong RBS-sfGFP (in pSB1C3)
Results
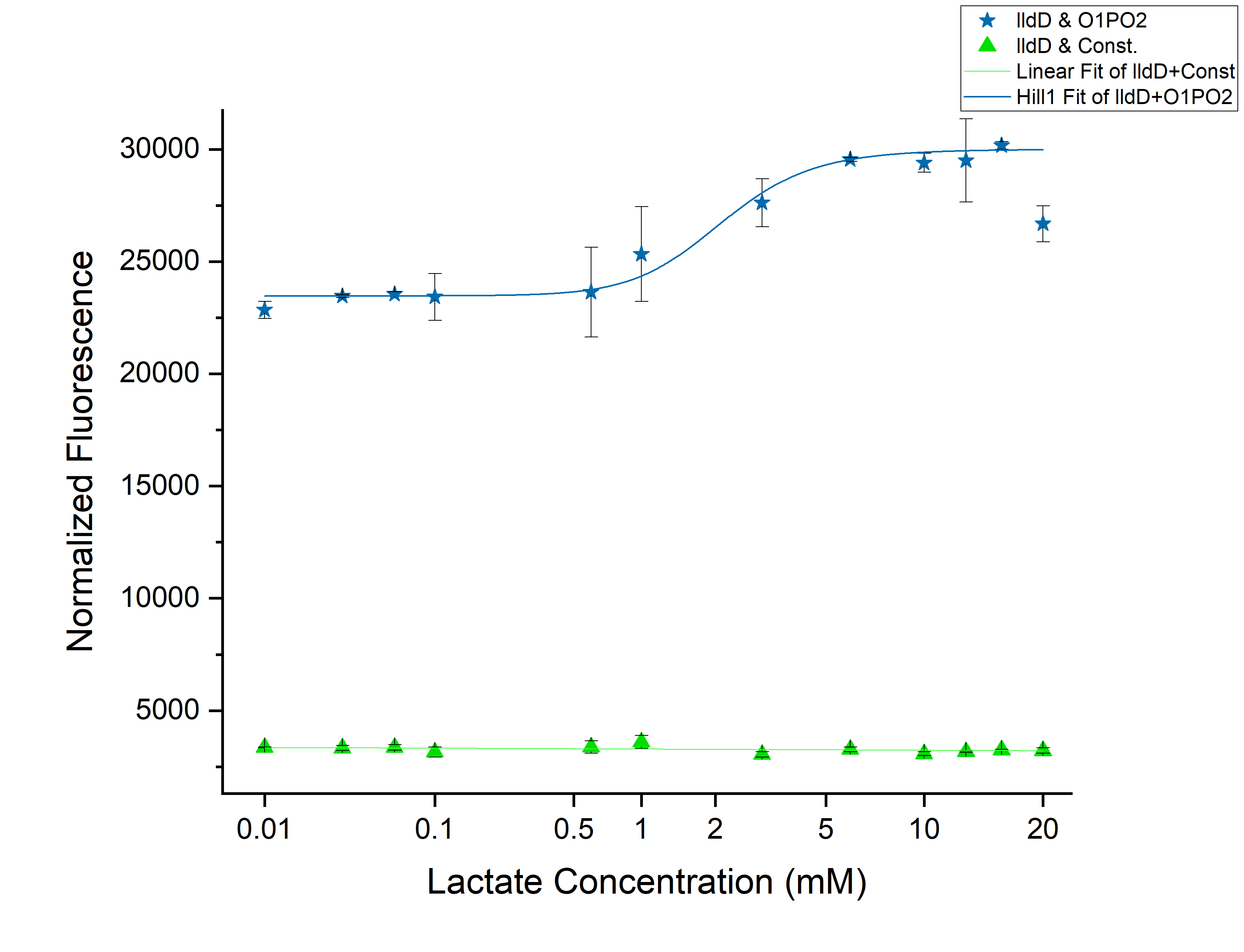
From the above
Comparison between only O1PO2 and O1PO2 along with lldD