Difference between revisions of "Part:BBa B0033"
| Line 6: | Line 6: | ||
===Improvement=== | ===Improvement=== | ||
<b>Group:iGEM2019_JiangnanU_China</b> | <b>Group:iGEM2019_JiangnanU_China</b> | ||
| − | |||
This year, we improved the sequence of the RBS <bbpart>BBa_B0033</bbpart>, and got the RBS <bbpart>BBa_K3137001</bbpart>. | This year, we improved the sequence of the RBS <bbpart>BBa_B0033</bbpart>, and got the RBS <bbpart>BBa_K3137001</bbpart>. | ||
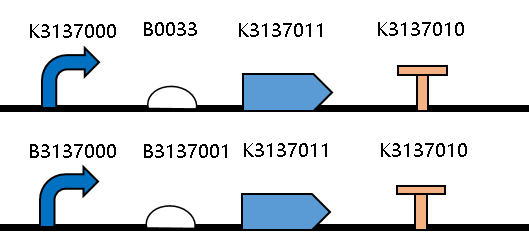
| − | [[Image:JiangnanUchina improve 1.png|400px|thumb|center|Figure1. | + | [[Image:JiangnanUchina improve 1.png|400px|thumb|center|Figure1. Maps of the constructed plasmids]] |
As we all know, RBS refers to a sputum-rich untranslated region upstream of the initiation codon AUG. <bbpart>BBa_B0033</bbpart> is a weaker RBS based on Ron Weiss thesis. Compared with <bbpart>BBa_B0030</bbpart>, <bbpart>BBa_B0031</bbpart>, <bbpart>BBa_B0032</bbpart>, <bbpart>BBa_B0034</bbpart>, <bbpart>BBa_B0033</bbpart> is weakest, and whose RBS strength is 0.35% of <bbpart>BBa_B0034</bbpart>. By analyzing their sequence, we found that <bbpart>BBa_B0033</bbpart> has fewer purines and no not have AAA sequence. SO we designed the RBS by adjusting the proportion of bases. | As we all know, RBS refers to a sputum-rich untranslated region upstream of the initiation codon AUG. <bbpart>BBa_B0033</bbpart> is a weaker RBS based on Ron Weiss thesis. Compared with <bbpart>BBa_B0030</bbpart>, <bbpart>BBa_B0031</bbpart>, <bbpart>BBa_B0032</bbpart>, <bbpart>BBa_B0034</bbpart>, <bbpart>BBa_B0033</bbpart> is weakest, and whose RBS strength is 0.35% of <bbpart>BBa_B0034</bbpart>. By analyzing their sequence, we found that <bbpart>BBa_B0033</bbpart> has fewer purines and no not have AAA sequence. SO we designed the RBS by adjusting the proportion of bases. | ||
| − | [[Image:JiangnanUchina improve 2.png|400px|center]] | + | [[Image:JiangnanUchina improve 2.png|400px|thumb|center|Figure2. Sequences of the original RBS and improved RBS]] |
| − | Then we ligated the two RBS with promotor rsmH (<bbpart>BBa_K3137000</bbpart>) and gene gfp (<bbpart>BBa_K3137011</bbpart>) and transform into E. coli BL21 to compare their green fluorescence. And take E. coli BL21 as a control. | + | Then we ligated the two RBS with promotor <i>rsmH</i> (<bbpart>BBa_K3137000</bbpart>) and gene <i>gfp</i> (<bbpart>BBa_K3137011</bbpart>) and transform into <i>E. coli</i> BL21 to compare their green fluorescence. And take <I>E. coli</i> BL21 as a control. |
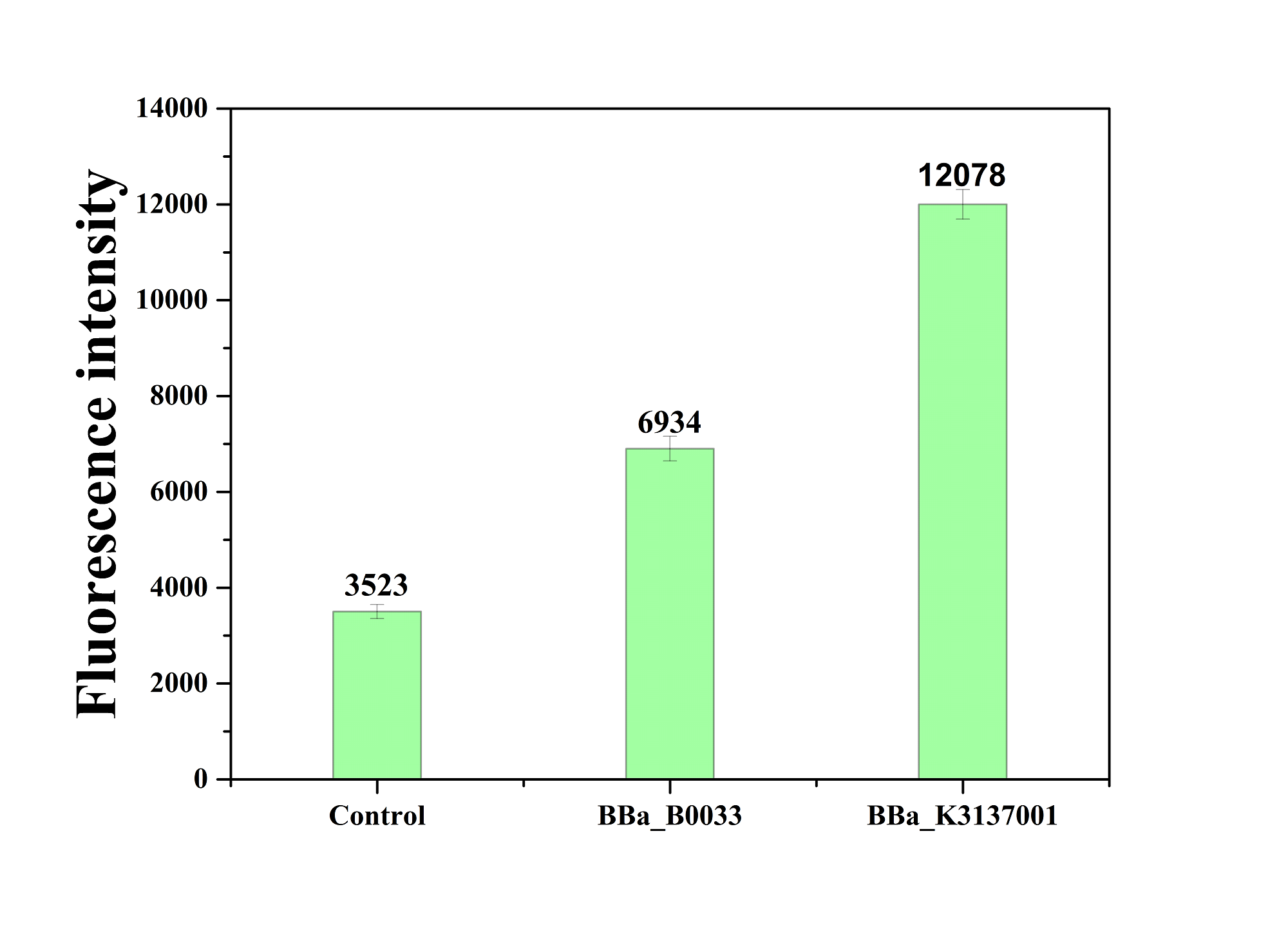
| − | The figure below shows that BBa_K3137001 has the stronger fluorescently protein expression than BBa_B0033, that means it’s efficient to increase the protein expression by increasing the number of purines in the RBS sequence, which is beneficial to the binding of ribosomes. | + | The figure below shows that BBa_K3137001 has the stronger fluorescently protein expression than <bbpart>BBa_B0033</bbpart>, that means it’s efficient to increase the protein expression by increasing the number of purines in the RBS sequence, which is beneficial to the binding of ribosomes. |
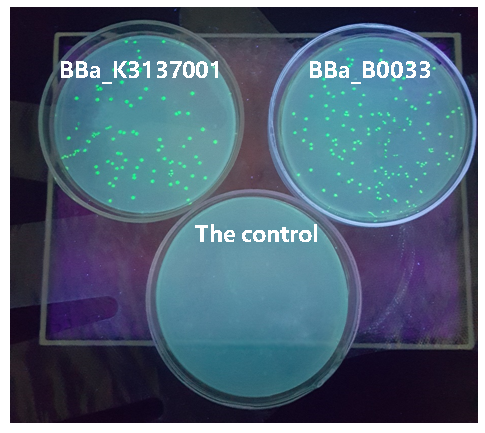
| − | [[Image:JiangnanUchina improve 3.png|400px|]] | + | [[Image:JiangnanUchina improve 3.png|400px|thumb|left|Figure3. Expression of <I>gfp</i> driven by <bbpart>BBa_K3137001</bbpart>, <bbpart>BBa_B0033</bbpart> and control]] |
| − | [[Image:JiangnanUchina improve 4.png|400px|]] | + | [[Image:JiangnanUchina improve 4.png|400px|thumb|right|Figure4. Fluorescence intensity of <i>gfp</i> driven by <bbpart>BBa_K3137001</bbpart>, <bbpart>BBa_B0033</bbpart> and control]] |
| + | |||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 22: | Line 25: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_B0033 SequenceAndFeatures</partinfo> | <partinfo>BBa_B0033 SequenceAndFeatures</partinfo> | ||
| + | |||
===Functional Parameters=== | ===Functional Parameters=== | ||
Revision as of 05:37, 21 October 2019
RBS.4 (weaker) -- derivative of BBa_0030
Weaker RBS based on Ron Weiss thesis. Strengths relative to BBa_B0030, BBa_B0031, BBa_B0032.
Improvement
Group:iGEM2019_JiangnanU_China This year, we improved the sequence of the RBS BBa_B0033, and got the RBS BBa_K3137001.
As we all know, RBS refers to a sputum-rich untranslated region upstream of the initiation codon AUG. BBa_B0033 is a weaker RBS based on Ron Weiss thesis. Compared with BBa_B0030, BBa_B0031, BBa_B0032, BBa_B0034, BBa_B0033 is weakest, and whose RBS strength is 0.35% of BBa_B0034. By analyzing their sequence, we found that BBa_B0033 has fewer purines and no not have AAA sequence. SO we designed the RBS by adjusting the proportion of bases.
Then we ligated the two RBS with promotor rsmH (BBa_K3137000) and gene gfp (BBa_K3137011) and transform into E. coli BL21 to compare their green fluorescence. And take E. coli BL21 as a control. The figure below shows that BBa_K3137001 has the stronger fluorescently protein expression than BBa_B0033, that means it’s efficient to increase the protein expression by increasing the number of purines in the RBS sequence, which is beneficial to the binding of ribosomes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| biology | -NA- |
| efficiency | 0.01 |
(Relative to BBa_B0034)