Difference between revisions of "Part:BBa K864402"
(→Contribution) |
(→Contribution) |
||
| Line 26: | Line 26: | ||
</ul> | </ul> | ||
<br><br> | <br><br> | ||
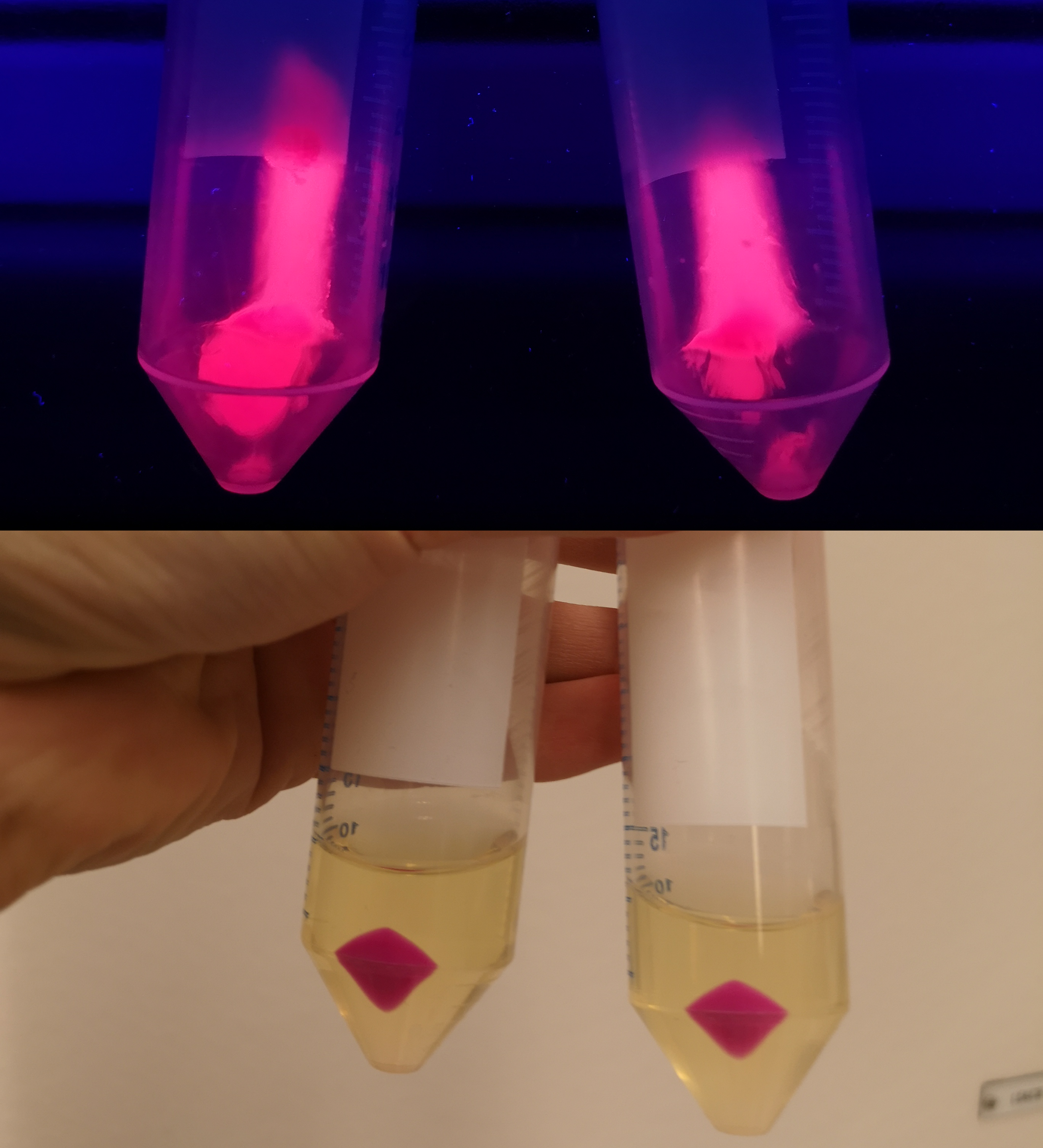
| − | <p> To verify eforReds absorbance and emission, the construct was expressed in E.coli BL21 (DE3). The bacteria containing the constitutive promotor (p.Cons) was compared to a negative control (<b>Figure 1</b>, left side). Thereafter the eforRed bacteria was centrifuged which displayed a pink pellet (<b>Figure 1</b>, top-right corner). To demonstrate the fluorescence of eforRed, the pellet was placed on a UV-table emitting a wavelength of 302 nm, (<b>Figure 1</b>, down-right corner), which exhibited a pink glowing colour.</p> | + | <p> To verify eforReds absorbance and emission, the construct was expressed in E.coli BL21 (DE3) gold cells. The bacteria containing the constitutive promotor (p.Cons) was compared to a negative control (<b>Figure 1</b>, left side). Thereafter the eforRed bacteria was centrifuged which displayed a pink pellet (<b>Figure 1</b>, top-right corner). To demonstrate the fluorescence of eforRed, the pellet was placed on a UV-table emitting a wavelength of 302 nm, (<b>Figure 1</b>, down-right corner), which exhibited a pink glowing colour.</p> |
<br><br> | <br><br> | ||
</html>[[Image:T--Linkoping_Sweden--pcons-eforred.jpeg|460px]]<html> | </html>[[Image:T--Linkoping_Sweden--pcons-eforred.jpeg|460px]]<html> | ||
Revision as of 11:49, 15 October 2019
J23110-B0034-eforRed
eforRed eforRed is previously described as BBa_K592012. We submitted a functionally active variant with J23110 and B0034.
Contribution
Group: Linkoping_Sweden iGEM 2019
Author: Andreas Holmqvist and Leo Juhlin
Summary:
In this contribution we characterized the visual absorbance and the fluorescence of this construct. We also tested the oxygen dependency of the chromophore protein expression in E.coli BL21(DE3) cells and then measured the absorbance of the cells with a plate reader.
Documentation:
- the BBa_B0034 ribosome binding site
- the BBa_J23110 Constitutive promotor
To verify eforReds absorbance and emission, the construct was expressed in E.coli BL21 (DE3) gold cells. The bacteria containing the constitutive promotor (p.Cons) was compared to a negative control (Figure 1, left side). Thereafter the eforRed bacteria was centrifuged which displayed a pink pellet (Figure 1, top-right corner). To demonstrate the fluorescence of eforRed, the pellet was placed on a UV-table emitting a wavelength of 302 nm, (Figure 1, down-right corner), which exhibited a pink glowing colour.

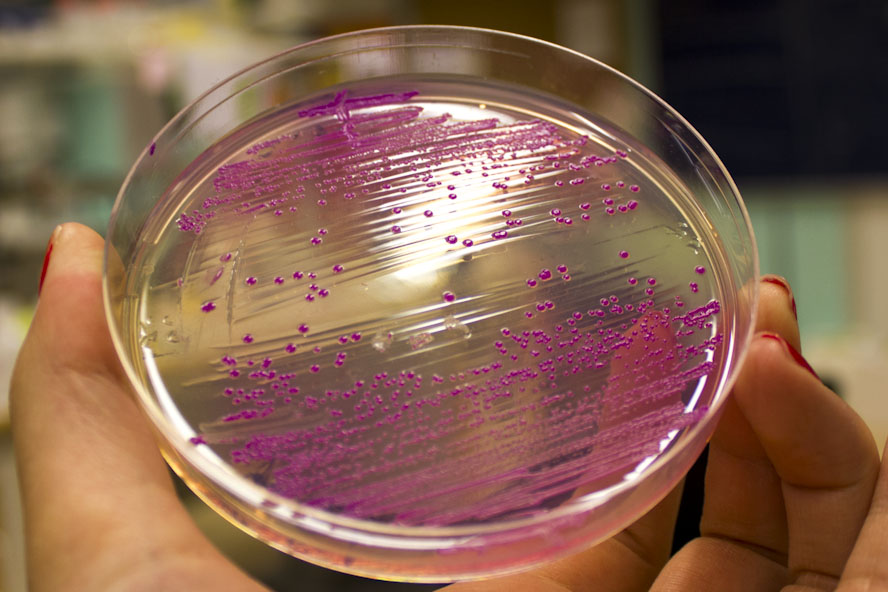
To test the fluorescence, E.coli BL21(DE3) containing pCons eforRed were spread on a agar plate containing 25 µg/ml chloramphenicol. This was done to illustrate the ability of eforRed to emit strong fluorescens on an agar plate in UV-light(SKRIVA MERA SOM EN METOD)??, and at the same time have a clear Burgundy color in visible light on the plate.
To examine the effect of oxygen supply on the recombinant protein production of eforRed in e.Coli BL21 (DE3) a platereading was conducted. The oxygen supply was varied by sticking different amount of holes in the plastic film of a 96-well plate. The experiment showed that the access to oxygen effects E.colis recombinant protein production of eforred in a 96 well plate.
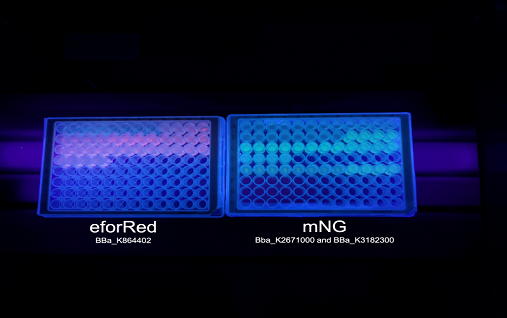
Figure 3. To the left is a spectrophotometric experiment where varying levels of oxygen supply were tested. The holes were made in the plastic cover of a 96-well plate and run for 16 hours in 37 degrees Celsius, this results shows the oxygen dependency of eforReds chromophore/fluorophore development. To the right is this biobrick in a 96-well plate, the culture has been in the well O.N in 37 degrees Celsius with varying levels of oxygen supply. The plate to the right is different expression systems used to test a strong green/yellow fluorescent protein. These results show eforReds ability to fluorescence, making it a fluorescent as well as a chromoprotein. Both plates were illuminated in 302 nm UV-light.
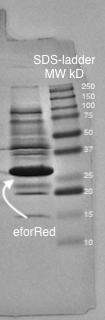
A study of the molecular weight of p.Cons-eforRed expressed in E.coli BL21(DE3) was done by sonicating the E.coli cells containing p.Cons-eforRed and then performing a SDS-PAGE Electrophoresis on the lysate (Figure z.).
Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Note: This part did not have a reference sequence. A reference sequence has since been added based on the part's documentation; a composite part using the following basic parts: No part name specified with partinfo tag. - BBa_B0034 - BBa_K592012- iGEM HQ