Difference between revisions of "Help:Plasmid backbones/Glossary"
| Line 2: | Line 2: | ||
__notoc__ __NOEDITSECTION__ | __notoc__ __NOEDITSECTION__ | ||
| − | + | ||
| − | + | ||
Here at the Registry, we use the term "plasmid" in three different ways: | Here at the Registry, we use the term "plasmid" in three different ways: | ||
| − | *" | + | *"'''[[Help:Plasmids/Construction Plasmids#Plasmid backbone|Plasmid backbone]]'''" - the vector alone, WITHOUT any BioBrick part |
| − | *" | + | *"'''[[Help:Plasmids/Construction Plasmids#Construction plasmid|Construction plasmid]]'''" - BioBrick WITH a default BioBrick part(s) to help you with [[Help:BioBrick Assembly|assembly]] |
| − | *" | + | *"'''[[Help:Plasmids/Construction Plasmids#Plasmid with any BioBrick part(s)|Plasmid with any BioBrick part(s)]]'''" - BioBrick vector WITH some BioBrick part(s). |
| − | | | + | |
| + | {| | ||
|colspan=2| | |colspan=2| | ||
==Plasmid backbone== | ==Plasmid backbone== | ||
|- | |- | ||
| − | | | + | | |
| BioBrick plasmids form the "backbone" of all BioBrick parts. Every BioBrick part is maintained on at least one BioBrick plasmid. Sometimes, a part is maintained on multiple BioBrick plasmid backbones, which vary in antibiotic resistance and replication origin (plasmid copy number). | | BioBrick plasmids form the "backbone" of all BioBrick parts. Every BioBrick part is maintained on at least one BioBrick plasmid. Sometimes, a part is maintained on multiple BioBrick plasmid backbones, which vary in antibiotic resistance and replication origin (plasmid copy number). | ||
| − | + | ||
| − | + | ||
*An example of a BioBrick plasmid without a part insert is '''[[Part:pSB1A3|pSB1A3]]'''. You can find more on plasmid naming [[Help:Plasmids/Nomenclature|here]] | *An example of a BioBrick plasmid without a part insert is '''[[Part:pSB1A3|pSB1A3]]'''. You can find more on plasmid naming [[Help:Plasmids/Nomenclature|here]] | ||
*To find a plasmid part using the "jump to part" sidebar function, erase the default "BBa_" prefix, and enter only the [[Help:Plasmids/Nomenclature|plasmid part name]] (e.g., pSB4K5 or pSB3K3 or pSB1AC3). | *To find a plasmid part using the "jump to part" sidebar function, erase the default "BBa_" prefix, and enter only the [[Help:Plasmids/Nomenclature|plasmid part name]] (e.g., pSB4K5 or pSB3K3 or pSB1AC3). | ||
|- | |- | ||
|colspan=2| | |colspan=2| | ||
| − | |||
==Construction plasmid== | ==Construction plasmid== | ||
|- | |- | ||
| − | | | + | |[[Image:BioBrickconstructionplasmid.png]] |
| − | [ | + | | If you just want one of the BioBrick plasmids, it is probably available in the iGEM DNA distribution with the part |
| − | | | + | |
| − | If you just want one of the BioBrick plasmids, it is probably available in the iGEM DNA distribution with the part | + | |
[[Part:BBa_P1010| BBa_P1010]], [[Part:BBa_I52001| BBa_I52001]] or [[Part:BBa_I52002| BBa_I52002]] in the cloning site. You will need the cell strain [[Part:BBa_V1005|DB3.1]] since each of these three parts constitutively expresses the protein CcdB which is toxic to most strains. | [[Part:BBa_P1010| BBa_P1010]], [[Part:BBa_I52001| BBa_I52001]] or [[Part:BBa_I52002| BBa_I52002]] in the cloning site. You will need the cell strain [[Part:BBa_V1005|DB3.1]] since each of these three parts constitutively expresses the protein CcdB which is toxic to most strains. | ||
| − | + | ||
| − | + | ||
When making a brand new BioBrick part, you will need to choose an appropriate BioBrick plasmid (with the right antibiotic resistance marker, copy number etc.) for your construction purposes. To make construction and assembly of BioBrick parts easier, most BioBrick plasmids include the ''ccdB'' positive selection marker (encoded in BioBrick parts [[Part:BBa_P1010| BBa_P1010]], [[Part:BBa_I52001| BBa_I52001]] and [[Part:BBa_I52002| BBa_I52002]]) by default. The ''ccdB'' positive selection marker produces a toxic protein which kills most ''E. coli'' strains. Successful insertion of a BioBrick part into a BioBrick vector replaces the ''ccdB'' gene and enables cells to grow and divide to form colonies. A common problem during BioBrick part construction and assembly is that BioBrick vectors are not 100% digested when cut with restriction enzymes. The ''ccdB'' toxic gene ensures that cells transformed with uncut or religated plasmids do not grow. Thus, any colonies you obtain after a BioBrick part construction or assembly are more likely to be correct. | When making a brand new BioBrick part, you will need to choose an appropriate BioBrick plasmid (with the right antibiotic resistance marker, copy number etc.) for your construction purposes. To make construction and assembly of BioBrick parts easier, most BioBrick plasmids include the ''ccdB'' positive selection marker (encoded in BioBrick parts [[Part:BBa_P1010| BBa_P1010]], [[Part:BBa_I52001| BBa_I52001]] and [[Part:BBa_I52002| BBa_I52002]]) by default. The ''ccdB'' positive selection marker produces a toxic protein which kills most ''E. coli'' strains. Successful insertion of a BioBrick part into a BioBrick vector replaces the ''ccdB'' gene and enables cells to grow and divide to form colonies. A common problem during BioBrick part construction and assembly is that BioBrick vectors are not 100% digested when cut with restriction enzymes. The ''ccdB'' toxic gene ensures that cells transformed with uncut or religated plasmids do not grow. Thus, any colonies you obtain after a BioBrick part construction or assembly are more likely to be correct. | ||
| − | *For a more in-depth walkthrough of how to use construction plasmids, check the [[Registry FAQ|FAQs]] or [[Featured Parts:Cell Death|"Cell Death" Featured Parts]] section | + | *For a more in-depth walkthrough of how to use construction plasmids, check the [[Registry FAQ|FAQs]] or [[Featured Parts:Cell Death|"Cell Death" Featured Parts]] section. |
|- | |- | ||
|colspan=2| | |colspan=2| | ||
Revision as of 22:10, 4 September 2008
Here at the Registry, we use the term "plasmid" in three different ways:
- "Plasmid backbone" - the vector alone, WITHOUT any BioBrick part
- "Construction plasmid" - BioBrick WITH a default BioBrick part(s) to help you with assembly
- "Plasmid with any BioBrick part(s)" - BioBrick vector WITH some BioBrick part(s).
Plasmid backbone | |
BioBrick plasmids form the "backbone" of all BioBrick parts. Every BioBrick part is maintained on at least one BioBrick plasmid. Sometimes, a part is maintained on multiple BioBrick plasmid backbones, which vary in antibiotic resistance and replication origin (plasmid copy number).
| |
Construction plasmid | |
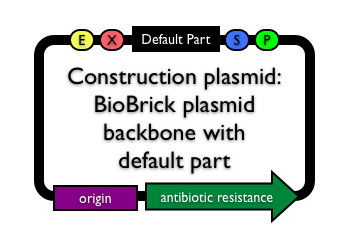
|
If you just want one of the BioBrick plasmids, it is probably available in the iGEM DNA distribution with the part
BBa_P1010, BBa_I52001 or BBa_I52002 in the cloning site. You will need the cell strain DB3.1 since each of these three parts constitutively expresses the protein CcdB which is toxic to most strains. When making a brand new BioBrick part, you will need to choose an appropriate BioBrick plasmid (with the right antibiotic resistance marker, copy number etc.) for your construction purposes. To make construction and assembly of BioBrick parts easier, most BioBrick plasmids include the ccdB positive selection marker (encoded in BioBrick parts BBa_P1010, BBa_I52001 and BBa_I52002) by default. The ccdB positive selection marker produces a toxic protein which kills most E. coli strains. Successful insertion of a BioBrick part into a BioBrick vector replaces the ccdB gene and enables cells to grow and divide to form colonies. A common problem during BioBrick part construction and assembly is that BioBrick vectors are not 100% digested when cut with restriction enzymes. The ccdB toxic gene ensures that cells transformed with uncut or religated plasmids do not grow. Thus, any colonies you obtain after a BioBrick part construction or assembly are more likely to be correct.
|
Plasmid with any BioBrick part(s) | |
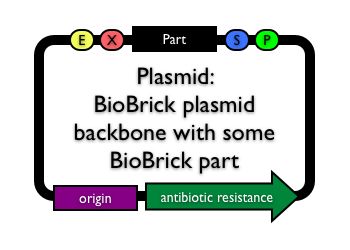
|
This term refers to a plasmid backbone with one or more BioBrick parts inserted. An example of this can be seen to the left, where a BioBrick part has been inserted into between the BioBrick prefix (denoted by E and X) and the BioBrick suffix (denoted by S and P). |
