Difference between revisions of "Part:BBa K2675040"
| Line 5: | Line 5: | ||
This part is the AimR receptor of the ''arbitrium'' peptide of the ''Bacillus'' phage phi3T [1] equipped with a synthetic RBS ([[Part:BBa_K2675010|BBa_K2675010]]) and placed under the control of the constitutive strong promoter [[Part:BBa_J23102|BBa_J23102]] and of the strong terminator ECK120029600 ([[Part:BBa_K2486006|BBa_K2486006]]). | This part is the AimR receptor of the ''arbitrium'' peptide of the ''Bacillus'' phage phi3T [1] equipped with a synthetic RBS ([[Part:BBa_K2675010|BBa_K2675010]]) and placed under the control of the constitutive strong promoter [[Part:BBa_J23102|BBa_J23102]] and of the strong terminator ECK120029600 ([[Part:BBa_K2486006|BBa_K2486006]]). | ||
| − | This part is functional in ''E. coli'': it expresses AimR that behaves as a transcriptional regulator. | + | This part is functional in ''E. coli'': it expresses AimR that behaves as a transcriptional regulator of sfGFP-LVAtag expression from [[Part:BBa_K2675058|BBa_K2675058]] and [[Part:BBa_K2675059|BBa_K2675059]]. |
It is an improvement of [[Part:BBa_K2279000|BBa_K2279000]]. | It is an improvement of [[Part:BBa_K2279000|BBa_K2279000]]. | ||
Latest revision as of 00:47, 18 October 2018
AimR of phage phi3T expression cassette
This part is the AimR receptor of the arbitrium peptide of the Bacillus phage phi3T [1] equipped with a synthetic RBS (BBa_K2675010) and placed under the control of the constitutive strong promoter BBa_J23102 and of the strong terminator ECK120029600 (BBa_K2486006).
This part is functional in E. coli: it expresses AimR that behaves as a transcriptional regulator of sfGFP-LVAtag expression from BBa_K2675058 and BBa_K2675059.
It is an improvement of BBa_K2279000.
Usage and Biology
The switch from lytic-to-lysogenic cycle of the Bacillus phage phi3T is based on the expression of a single transcript, AimX [1]. The expression of AimX is controlled by AimR (BBa_K2279000) who was described to be a transcriptional regulator of the pAimX promoter in B. subtilis [1]. The exact binding site of AimR to DNA was not precisely described, but it was mapped by ChIPseq in the intergenic region between AimP and AimX genes, a.k.a. the pAimX(full) promoter (BBa_K2675020). We bring experimental evidence that the AimR binding site is located in the pAimX(3) putative promoter a.k.a. the pAimX(short) promoter (BBa_K2675021).
pAimX(full) (BBa_K2675020) and pAimX(short) (BBa_K2675021) sequences, as well as their hybrid versions (BBa_K2675022, BBa_K2675023, BBa_K2675024, BBa_K2675025), proved not to be active promoters in E. coli. However, assuming that AimR retains its binding ability to its specific target sequence in E. coli, we considered converting the pAimX into an operator for regulation of an E. coli promoter. To do this, we needed to place a constitutive E. coli promoter upstream of pAimX and a reporter gene downstream of pAimX. In this design, transcription will be initiated at the upstream constitutive E. coli promoter and, in the absence of AimR, the RNA polymerase should be able to transcribe the pAimX region, the RBS and the reporter sequence up to the terminator. In the presence of AimR, AimR will bind to its specific recognition sequence mapped in pAimX(full) region and thus it should prevent the RNA polymerase from continuing the transcription. The expected results are: - in the absence of AimR: expression of the reporter gene - in the presence of AimR: no expression of the reporter gene Thus, AimR will be converted from an activator in Bacillus to a repressor in E. coli.
We chose the BBa_J23110 constitutive promoter from the Anderson Library that is reported to have a medium activity compared to the other members of this promoter family. A medium strength promoter allows more easily to detect variation in its activity (inhibition or activation) than a low or high strength promoter.
Then we placed the pAimX(full) and pAimX(short) sequences (BBa_K2675020 and BBa_K2675021) between BBa_J23110 and sfGFP-LVAtag (BBa_K2675006), and we thus constructed 2 composite parts: BBa_K2675057 and BBa_K2675058 respectively. During the cloning process, we selected a colony that, upon sequencing, revealed to have a truncated version of the J23110 with only the -10 box (BBa_K2675026) upstream of pAimX(short). As this colony was green under blue light (with amber filter) due to sfGFP expression, we continued characterising this cloning artefact for its interesting promoter activity (BBa_K2675059).
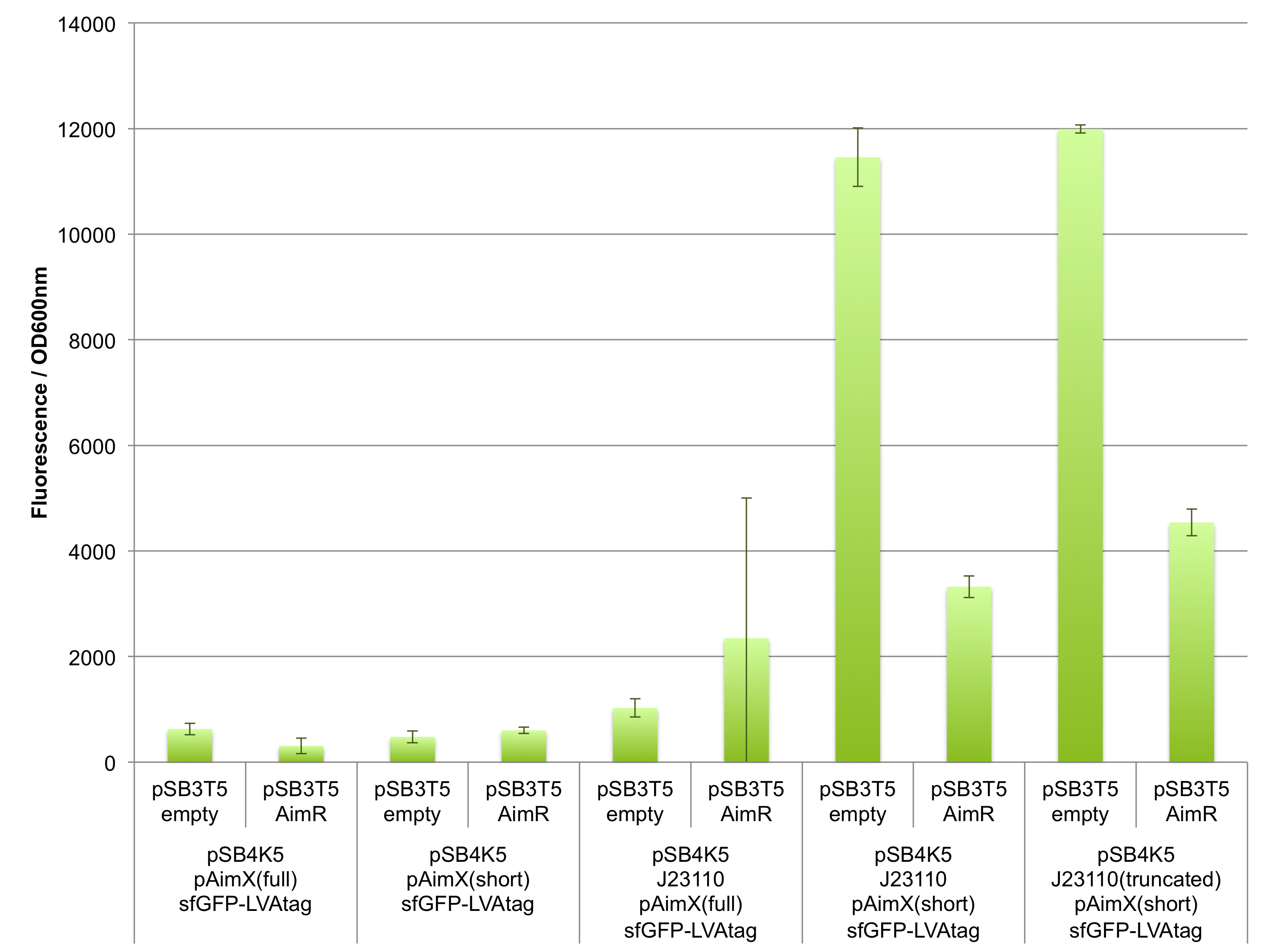
The results presented in Figures 1 and 2 show that the pAimX(short), but not pAimX(full) acts as expected as an operator for J23110 promoter in E. coli. Indeed, we can observe the fluorescent sfGFP expressed when this reporter gene is placed downstream of the composite promoter J23110 + pAimX(short) (BBa_K2675058). Moreover, the fluorescence decreases in the presence of AimR suggesting that AimR acts as predicted for this design: as a repressor. The same trend is observed also when the truncated version of J23110 in present upstream of pAimX(short) (BBa_K2675059).
However, no significant sfGFP expression was observed when the pAimX(full) sequence was inserted between between the J23110 promoter and sfGFP (BBa_K2675059). One plausible explanation is that transcription is initiated from the upstream J23110 promoter, but it ends prematurely at an internal terminator predicted to exist in the pAimX(full) sequence (see arrows marking a hairpin loop in Figure 1). In addition to proving that AimR can act as an effective repressor in E. coli, these experiments help identify the precise site of AimR binding to DNA: the pAimX(short) promoter (BBa_K2675021).
Figure 1: In vivo characterisation of sfGFP expression by E. coli cells harbouring the expression cassettes of sfGFP-LVAtag under the control of pAimX(full) promoter (BBa_K2675050), pAimX(short) promoter (BBa_K2675051), J23110 + pAimX(full) promoter (BBa_K2675057), J23110 + pAimX(short) promoter (BBa_K2675058), J23110 truncated promoter (-10 only) + pAimX(short) promoter (BBa_K2675059) in pSB4K5 backbone. The AimR was expressed as BBa_K2675040 on pSB3T5. An empty pSB3T5 served as negative control. Fluorescence values were normalised by OD600nm.
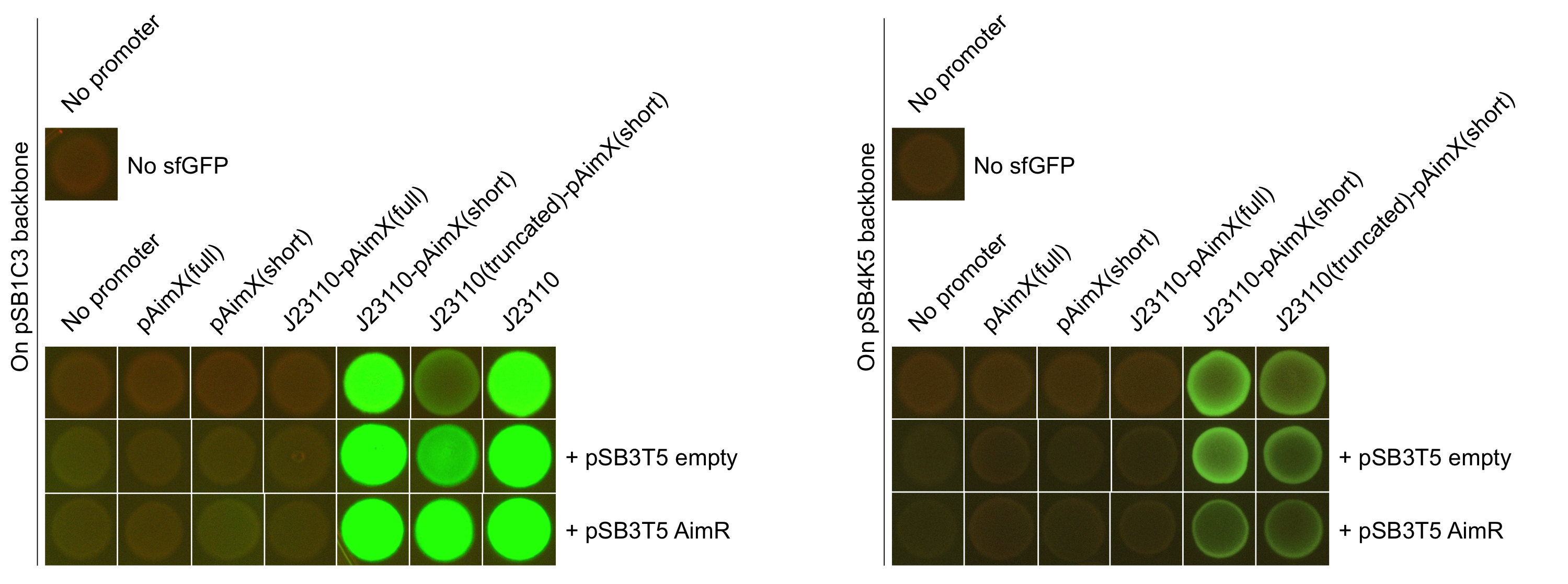
Figure 2: Pictures of E. coli cells harbouring the expression cassettes of sfGFP-LVAtag under the control of pAimX(full) promoter (BBa_K2675050), pAimX(short) promoter (BBa_K2675051), J23110 + pAimX(full) promoter (BBa_K2675057), J23110 + pAimX(short) promoter (BBa_K2675058), J23110 truncated promoter (-10 only) + pAimX(short) promoter (BBa_K2675059) in pSB1C3 or pSB4K5 backbones. The negative control has been performed with an empty pSB1C3 or pSB3K4 and the positive one with BBa_K2675056.
In Bacillus cells, AimR is inactivated upon binding of the ‘arbitrium’ hexapeptide SAIRGA [1]. To test if the repressor activity of AimR in E. coli is inhibited in the presence of the SAIRGA hexapeptide, we added in the culturing media increasing concentrations of the peptide. We can see on the graph presented in Figure 3 that fluorescence intensity and SAIRGA concentration are not really correlated since increasing concentration of SAIRGA doesn’t influence fluorescence output, neither in the absence nor in the presence of AimR. Although the fold-repression (the ratio between the Fluorescence/OD600nm values of the samples without AimR over those with AimR) increases slightly with increasing SAIRGA concentration, the variance of the fluorescence data are too high to draw a clear conclusion (Figures 4).
Instability of the SAIRGA in the medium or its inefficient import into E. coli may be other reasons why its effect on AimR is not obvious. The import of peptides in E. coli is dependent on the expression of transmembrane transporters in the cell plasma membrane. Considering that SAIRGA is a peptide signal, its import may be realised through the oligopeptide permease system (Opp) that is an ABC transporter capable of importing large oligopeptides into bacteria without selection for size, composition, sequence or charge [2]. The Opp system can be induced by supplementing the media with L-Leucine [2]. So, we decided to test the effect of SAIRGA peptide in culturing media supplemented with this amino acid. We also tested the effect of CaCl2, know to have a permeabilizing effect on E. coli plasma membrane. The results presented in Figure 5 are encouraging : the repression fold (the ratio between the Fluorescence/OD600nm values of the samples without AimR over those with AimR) is slightly increasing with increasing SAIRGA concentration.
Figure 3: In vivo characterization of sfGFP expression by E. coli cells harbouring the expression cassettes of sfGFP-LVAtag under the control of J23110 + pAimX(short) promoter (BBa_K2675058) and J23110 truncated promoter (-10 only) + pAimX(short) promoter (BBa_K2675059) in pSB4K5 backbone in the presence of increasing concentrations of SAIRGA hexapeptide in the culturing medium. The AimR was expressed as BBa_K2675040 on pSB3T5. An empty pSB3T5 served as negative control. Fluorescence values were normalised by OD600nm.
Figure 4: In vivo characterization of sfGFP expression by E. coli cells harbouring the expression cassettes of sfGFP-LVAtag under the control of J23110 + pAimX(short) promoter (BBa_K2675058) and J23110 truncated promoter (-10 only) + pAimX(short) promoter (BBa_K2675059) in pSB4K5 backbone in the presence of increasing concentrations of SAIRGA hexapeptide in the culturing medium. The AimR was expressed as BBa_K2675040 on pSB3T5. An empty pSB3T5 served as negative control. The results are present as the ratio between the Fluorescence/OD600nm values of the samples without AimR over those with AimR (represion fold).
Figure 5: In vivo characterization of sfGFP expression by E. coli cells harbouring the expression cassettes of sfGFP-LVAtag under the control of J23110 + pAimX(short) promoter (BBa_K2675058) and J23110 truncated promoter (-10 only) + pAimX(short) promoter (BBa_K2675059) in pSB4K5 backbone in the presence of increasing concentrations of SAIRGA hexapeptide in the culturing medium supplemented or not with 1 mM L-Leucine or 0.1 mM of CaCl2. The AimR was expressed as BBa_K2675040 on pSB3T5. An empty pSB3T5 served as negative control. The results are present as the ratio between the Fluorescence/OD600nm values of the samples without AimR over those with AimR (represion fold).
As the effect of externally added SAIRGA peptide was not very convincing, we tried expressing the SAIRGA peptide directly inside the cell. The results presented in Figure 6 do not show any effect of the co-expression, most probably because the SAIRGA peptide is expressed with specific secretion and protease-cleavage tags that remain unprocessed inside the cell.
Figure 6: In vivo characterization of sfGFP expression by E. coli cells harbouring the expression cassette of sfGFP-LVAtag under the control of J23110 + pAimX(short) promoter (BBa_K2675058) in pSB4K5 backbone in the presence of various SAIRGA expression plasmids (BBa_K2675041, BBa_K2675042, BBa_K2675043 and BBa_K2675044) in pSB4K5 backbone. The AimR was expressed as BBa_K2675040 on pSB3T5. Empty pSB1C3, pSB3T5 and pSB4K5 served as negative controls. Fluorescence values were normalised by OD600nm.
We have successfully transferred the ‘Arbitrium’ communication system from a Bacillus phage to E. coli. While doing so, we have engineered new “composite” promoters that convert the transcriptional activator AimR from Bacillus into a repressor in E. coli. We also have promising data that indicates conditional regulation of AimR repressor activity by the small signalling peptide SAIRGA.
References
[1] Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R. Communication between viruses guides lysis-lysogeny decisions. Nature (2017) 541, 488-493.
[2] Maio A, Brandi L, Donadio S, Gualerzi CO. The Oligopeptide Permease Opp Mediates Illicit Transport of the Bacterial P-site Decoding Inhibitor GE81112. Antibiotics (Basel) (2016) 5, 17.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]