Difference between revisions of "Part:BBa K2656013"
| Line 5: | Line 5: | ||
Part [https://parts.igem.org/Part:BBa_I746916 BBa_I746916] standarized as Phytobrick. Super Folder Green Fluorescent Protein coding sequence. It can be used to assemble transcriptional units with the Golden Gate assembly method, and also, it can be used as a Biobrick. | Part [https://parts.igem.org/Part:BBa_I746916 BBa_I746916] standarized as Phytobrick. Super Folder Green Fluorescent Protein coding sequence. It can be used to assemble transcriptional units with the Golden Gate assembly method, and also, it can be used as a Biobrick. | ||
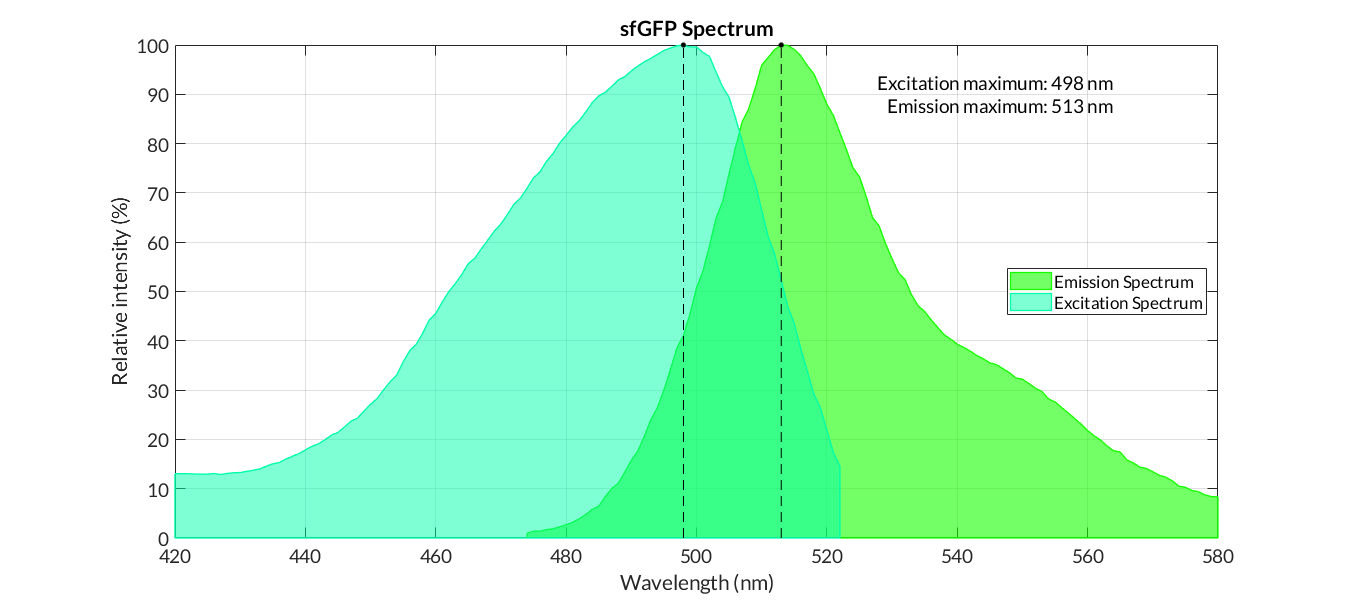
| − | In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we have assembled with it, we have obtained the emission and excitation spectra in the conditions of our equipment. To do this, we have used the transcriptional unit [https://parts.igem.org/Part:BBa_K2656100 BBa_K2656100 ] assembled in a Golden Braid alpha1 plasmid, with the replication origin pMB1. By using this protocol with the parameters of table 1, figure 1 has been obtained. | + | In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we have assembled with it, we have obtained the emission and excitation spectra in the conditions of our equipment. To do this, we have used the transcriptional unit [https://parts.igem.org/Part:BBa_K2656100 BBa_K2656100 ] assembled in a Golden Braid alpha1 plasmid'''(REF)''', with the replication origin pMB1. By using this protocol '''(REF)''' with the parameters of table 1, figure 1 has been obtained. |
Revision as of 19:53, 28 September 2018
sfGFP Coding Sequence
Part BBa_I746916 standarized as Phytobrick. Super Folder Green Fluorescent Protein coding sequence. It can be used to assemble transcriptional units with the Golden Gate assembly method, and also, it can be used as a Biobrick.
In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we have assembled with it, we have obtained the emission and excitation spectra in the conditions of our equipment. To do this, we have used the transcriptional unit BBa_K2656100 assembled in a Golden Braid alpha1 plasmid(REF), with the replication origin pMB1. By using this protocol (REF) with the parameters of table 1, figure 1 has been obtained.
| Parameter | Value | ||
| Sample number | 0 | ||
| Minimum excitation wavelenght (nm) | 0 | ||
| Maximum excitation wavelenght (nm) | 0 | ||
| Emission wavelenght (nm) | 0 | ||
| Minimum emission wavelenght (nm) | 0 | ||
| Maximum emission wavelenght (nm) | 0 | ||
| Excitation wavelenght (nm) | 0 | ||
| Gain (G) | 0 | ||
| Table 1. Parameters used to obtain the spectra | |||
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 14