Difference between revisions of "Part:BBa K112806"
(→Team NYMU-Taipei 2017: Successfully construct T4 endolysin into suicide mechanism) |
DanielaL19 (Talk | contribs) |
||
| Line 5: | Line 5: | ||
The lysozyme from enterobacteria phage T4 degrades peptidoglycan layer. | The lysozyme from enterobacteria phage T4 degrades peptidoglycan layer. | ||
| − | Group: (Michigan 2017) Author: (Aaron Renberg) Summary: We improved part BBa_K112806 from UC Berkeley’s 2008 project by optimizing the codons for translation in E. coli using IDT’s codon optimization tool making it much easier for future iGEM teams to use. The changes we made were T508C and A511G. Link: https://parts.igem.org/Part:BBa_K2301001 | + | Group: (Michigan 2017) Author: (Aaron Renberg) Summary: We improved part BBa_K112806 from UC Berkeley’s 2008 project by optimizing the codons for translation in E. coli using IDT’s codon optimization tool making it much easier for future iGEM teams to use. The changes we made were T508C and A511G. Additionally, we constructed three different versions (of varying promoter strength) of a temperature controlled kill switch using holin, endolysin and antiholin. Link: https://parts.igem.org/Part:BBa_K2301001 |
Revision as of 23:19, 1 November 2017
[T4 endolysin]
The lysozyme from enterobacteria phage T4 degrades peptidoglycan layer.
Group: (Michigan 2017) Author: (Aaron Renberg) Summary: We improved part BBa_K112806 from UC Berkeley’s 2008 project by optimizing the codons for translation in E. coli using IDT’s codon optimization tool making it much easier for future iGEM teams to use. The changes we made were T508C and A511G. Additionally, we constructed three different versions (of varying promoter strength) of a temperature controlled kill switch using holin, endolysin and antiholin. Link: https://parts.igem.org/Part:BBa_K2301001
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 271
Illegal AgeI site found at 341 - 1000COMPATIBLE WITH RFC[1000]
Team NYMU-Taipei 2017: Successfully construct T4 endolysin into suicide mechanism
Improvement
We successfully combine T4 endolysin with a constitutive promoter (BBa_J23106), a ribosome binding site (BBa_B0034) and a double terminator (BBa_B0010 and BBa_B0012). Furthermore, we construct T4 holin and T4 endolysin together as a functional suicide mechanism. Besides, in NYMU-Taipei 2017 team’s project, we also put the suicide mechanism into practice by constructing them with NrtA.
Result
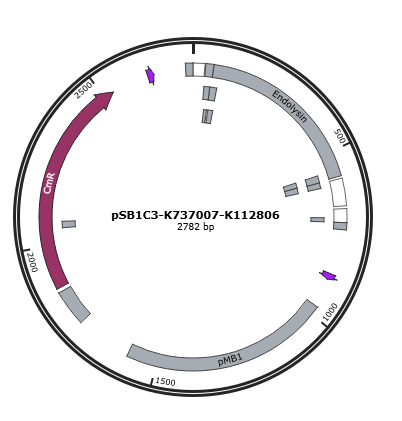
Figure 1 is the gene map of T4 endolysin construct, pSB1C3-K737007-K112806.
Figure 1
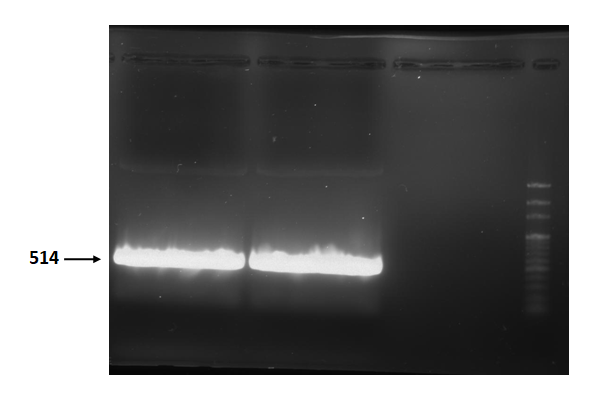
Figure 2 is electrophoresis result of endolysin (from BBa_K112806) PCR product.
The marker is 100bp. The length is 514bp as expected.
Figure 2
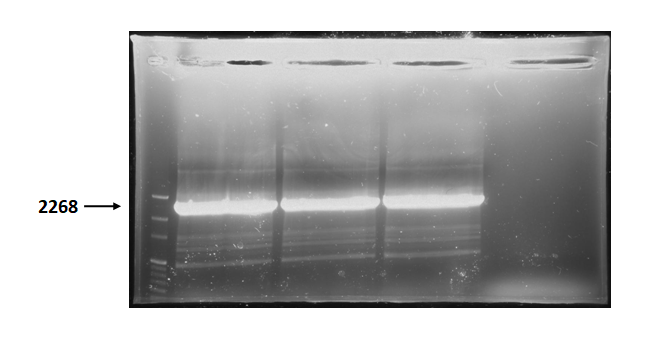
Figure 3 is electrophoresis result of endolysin backbone (from BBa_K737007) PCR product.
The marker is 100bp. The length is 2268bp as expected.
Figure 3
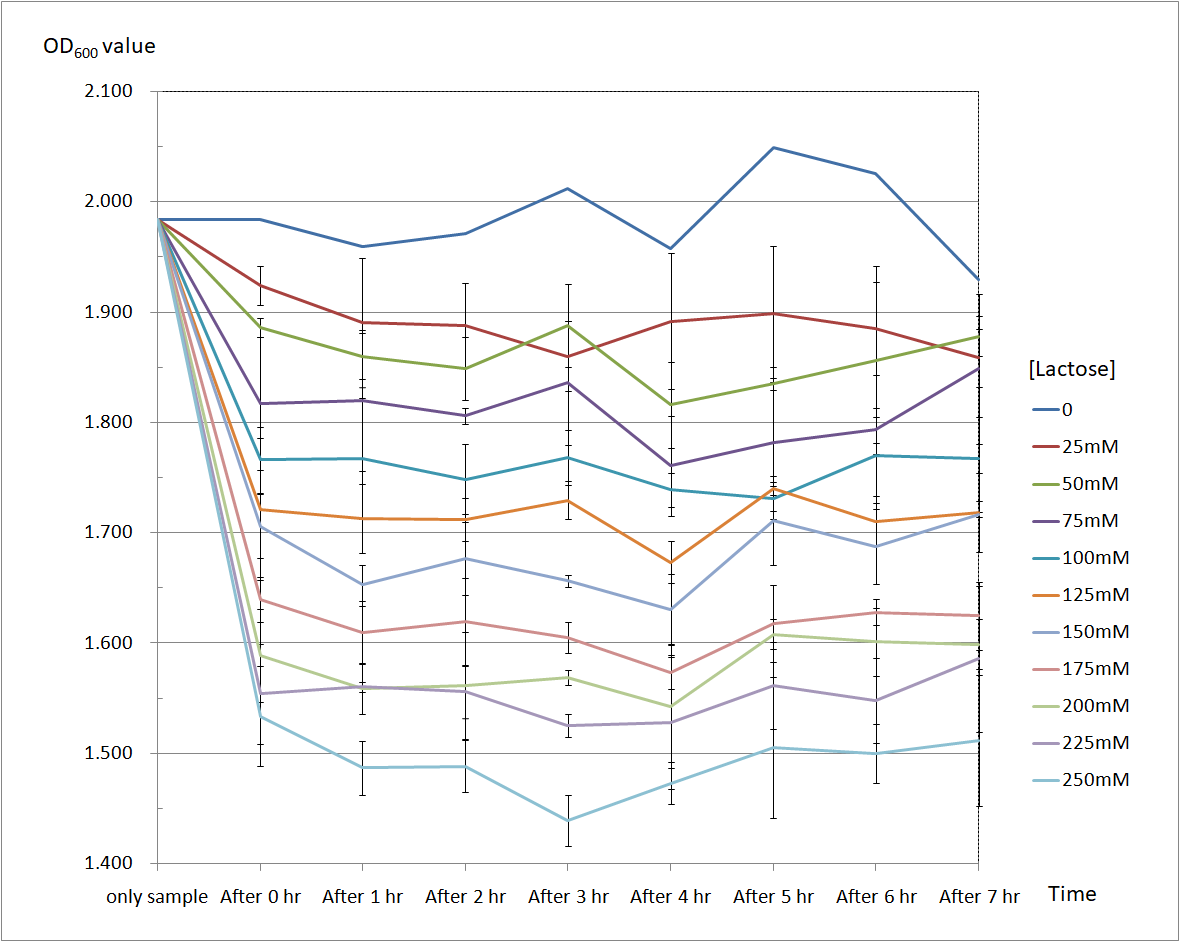
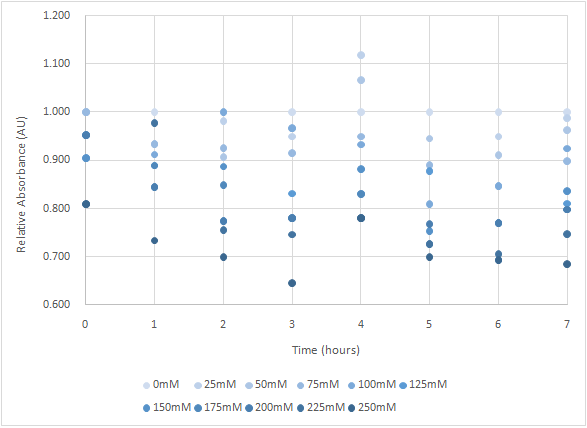
Figure 4 and Figure 5 are the results of suicide mechanism functional test.
Both figure show that our suicide mechanism can be induced by adding lactose, and the effectiveness of suicide mechanism goes better as the concentration of lactose goes higher.
Figure 4 is the bacterium with holin-endolysin construct.
As figure 3 shows, the suicide mechanism is induced immediately when lactose is added into the samples. Besides, we can see that the lactose concentration and the OD value of the bacterium with holin-endolysin construct are positively correlated, which means our suicide mechanism does work.
Figure 4
Figure 5 is the bacterium with holin-endolysin-NrtA construct.
As figure 4 shows, the trend of the relative absorbance is downward as the lactose is added to induce the suicide mechanism. The concentration of lactose is also positively correlated with the declining degree of relative absorbance.
Figure 5