Difference between revisions of "Part:BBa K2315034"
| Line 25: | Line 25: | ||
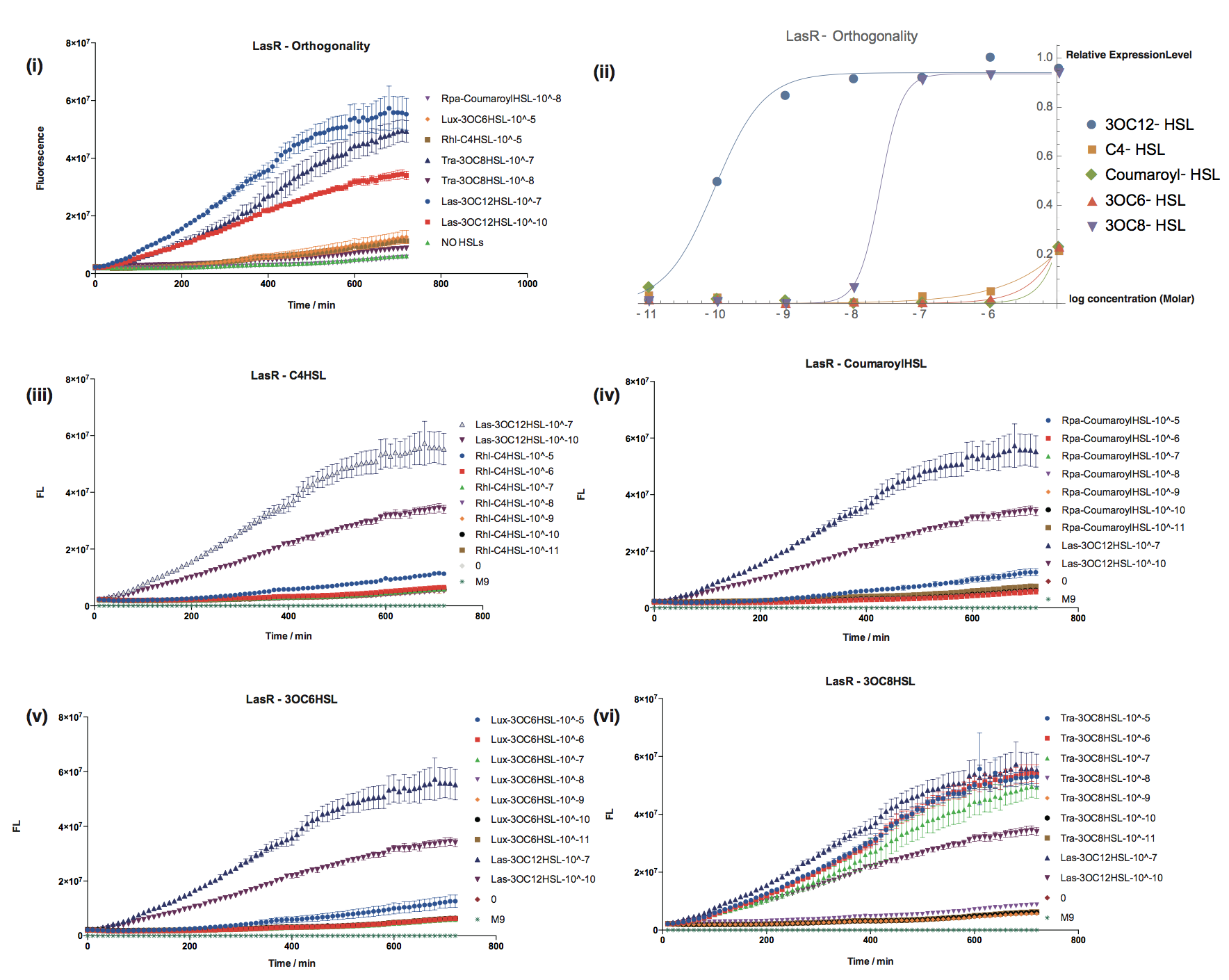
We have characterized crosstalk response of LasR to several non-cognate AHLs: | We have characterized crosstalk response of LasR to several non-cognate AHLs: | ||
[[File:T--Shanghaitech--LasRorthogonality.png|thumb|center|800px|<b>Fig. 2 Orthogonality test of LasR-pLas-GFP </b> (i):Fluorescent response to cognate and non-cognate AHLs (ii)Dose-Response curves for cognate and non-cognate AHLs (iii-vi)Fluorescent response to non-cognate AHLs in compared with 3OC12-HSL ]] | [[File:T--Shanghaitech--LasRorthogonality.png|thumb|center|800px|<b>Fig. 2 Orthogonality test of LasR-pLas-GFP </b> (i):Fluorescent response to cognate and non-cognate AHLs (ii)Dose-Response curves for cognate and non-cognate AHLs (iii-vi)Fluorescent response to non-cognate AHLs in compared with 3OC12-HSL ]] | ||
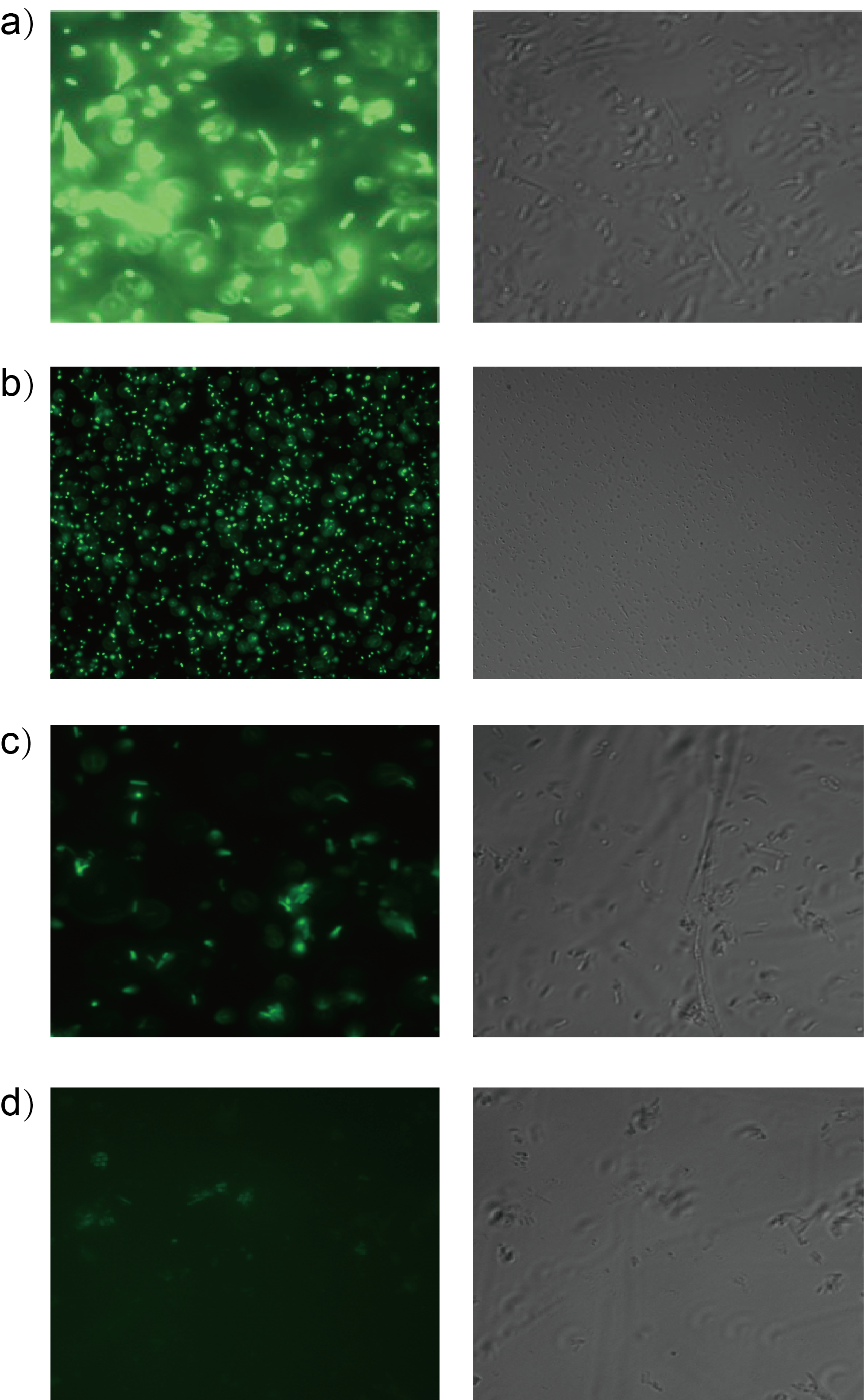
| − | Furthermore, we test it under fluorescence microscope. Fig. 3 shows four testing samples. a) Interlab Study Test Device 1 <bbpart>BBa_J364000</bbpart> b) Las molecule 3OC12-HSL as inducer c) Tra molecule 3OC8-HSL as inducer d) Rpa molecule Coumaroyl-HSL as inducer | + | Furthermore, we test it under fluorescence microscope. Fig. 3 shows four testing samples.<b> a) Interlab Study Test Device 1 <bbpart>BBa_J364000</bbpart> b) Las molecule 3OC12-HSL as inducer c) Tra molecule 3OC8-HSL as inducer d) Rpa molecule Coumaroyl-HSL as inducer</b> |
| + | [[File:T--Shanghaitech--2315034-3.png|thumb|center|700px|<b>Fig. 3 Orthogonality test under fluorescence microscope</b>]] | ||
It has shown that LasR is sensitive to it's cognate HSL and has obvious crosstalk with 3OC8-HSL in Tra System in relatively high concentration. | It has shown that LasR is sensitive to it's cognate HSL and has obvious crosstalk with 3OC8-HSL in Tra System in relatively high concentration. | ||
Revision as of 16:18, 1 November 2017
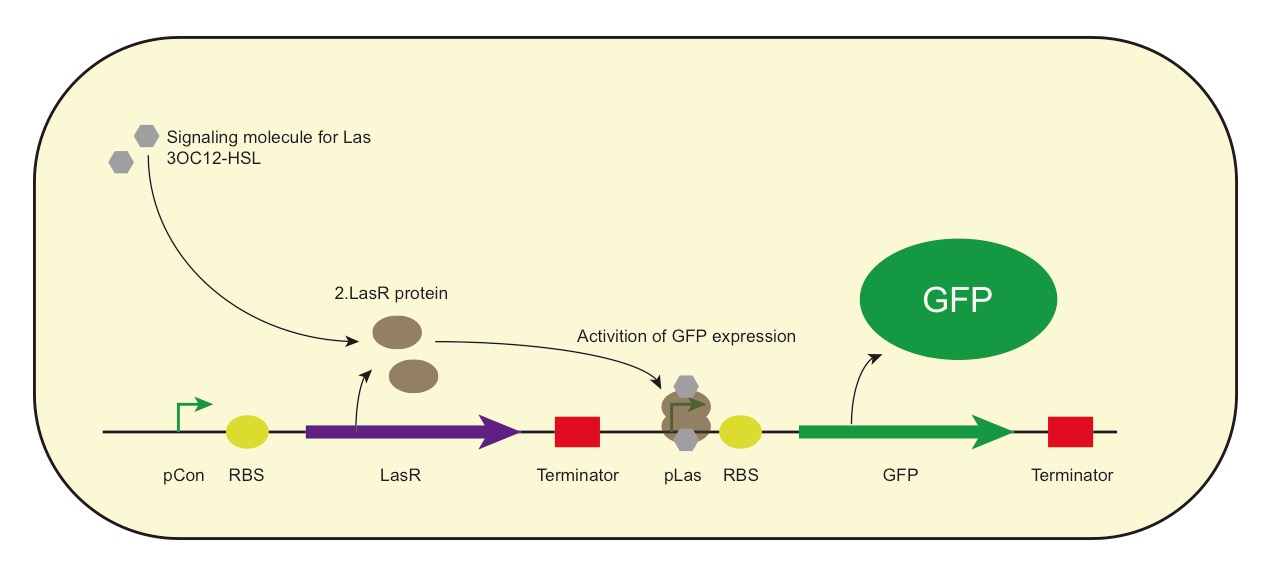
LasR-pLas-GFP HSL inducible fluorescent actuator
Group: Shanghaitech iGEM 2017
The AHL receiver LasR from P.aeruginosa activates expression of GFP protein in response to 3OC12-HSL.
This Receiver can be easily replaced by other AHL receivers in our collection. A full collection could be found in: http://2017.igem.org/Team:Shanghaitech/Library
Usage and Biology
When AHL is added with concentration higher than a critical value, the constitutively expressed LasR will bind to the AHL molecule 3OC12-HSL, dimerize and bind to the pLas regulatory sequence to activate GFP expression.
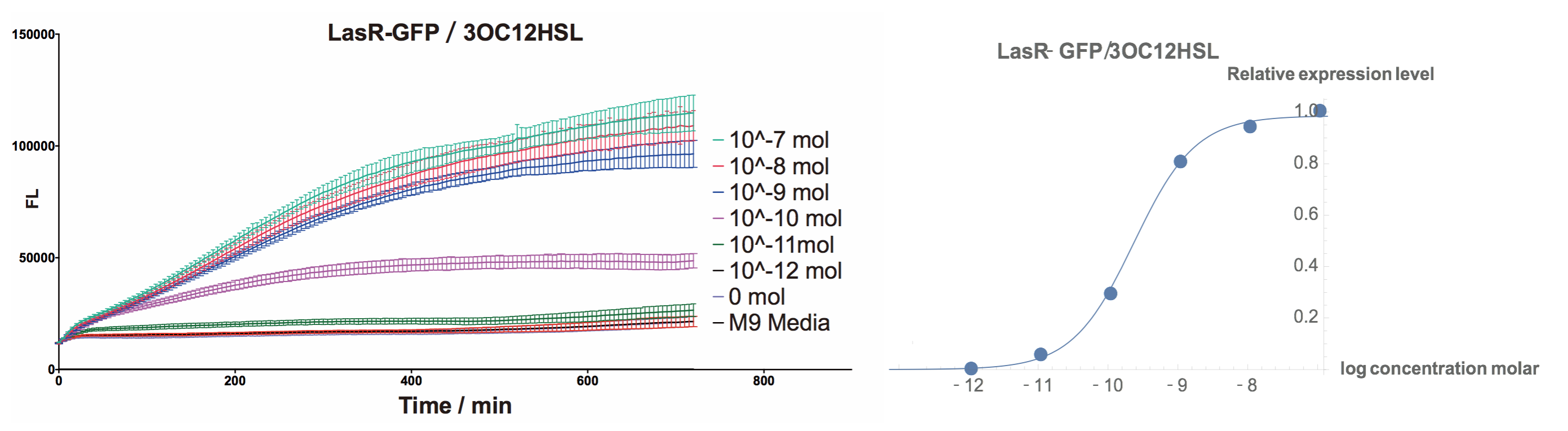
Fluorescent Response to cognate 3OC12-HSL
To test this part, we used standard 3OC12HSL (HSL produced by LasI in P.aeruginosa) to determine the response curve.
Orthogonality test against non-cognate inducers
We have characterized crosstalk response of LasR to several non-cognate AHLs:
Furthermore, we test it under fluorescence microscope. Fig. 3 shows four testing samples. a) Interlab Study Test Device 1 BBa_J364000 b) Las molecule 3OC12-HSL as inducer c) Tra molecule 3OC8-HSL as inducer d) Rpa molecule Coumaroyl-HSL as inducer
It has shown that LasR is sensitive to it's cognate HSL and has obvious crosstalk with 3OC8-HSL in Tra System in relatively high concentration.
Usages in our Project
We developed a measurement protocol using the fluorescent protein coupled AHL receiver germs to measure the actual AHLs concentration in high precision and sensitivity in compared with LC-MS.
AHL receiver from P.aeruginosa, actives expression of GFP protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30
Illegal NheI site found at 979 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 375
Illegal AgeI site found at 572 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1740