Difference between revisions of "Part:BBa K592012"
Georgedashen (Talk | contribs) (→Usage and Biology) |
Georgedashen (Talk | contribs) (→Usage and Biology) |
||
| Line 17: | Line 17: | ||
'''iGEM17_SJTU_BioX_Shanghai:'''<br> | '''iGEM17_SJTU_BioX_Shanghai:'''<br> | ||
| − | '''J23119+Target3+eforRed '''<partinfo>BBa_K2285012</partinfo> '''J23119+Target1+eforRed ''' <partinfo>BBa_K2285016</partinfo><br> | + | '''J23119+Target3+eforRed '''<partinfo>BBa_K2285012</partinfo><br> |
| + | '''J23119+Target1+eforRed ''' <partinfo>BBa_K2285016</partinfo><br> | ||
| + | |||
An improved part has been constructed. Since this part is a coding sequence, we added a RBS which is on the upstream of sfGFP <partinfo>BBa_K515005</partinfo> and terminator <partinfo>BBa_B1006</partinfo> by PCR. What's more, our device have a stem-loop structure(called Target) following the constitutive promoter J23119, to achieve further control of eforRed expression. | An improved part has been constructed. Since this part is a coding sequence, we added a RBS which is on the upstream of sfGFP <partinfo>BBa_K515005</partinfo> and terminator <partinfo>BBa_B1006</partinfo> by PCR. What's more, our device have a stem-loop structure(called Target) following the constitutive promoter J23119, to achieve further control of eforRed expression. | ||
Revision as of 01:32, 28 October 2017
eforRed, red chromoprotein
This chromoprotein eforRed naturally exhibits red/pink color when expressed. The color is slightly weaker than RFP. On agar plates and in liquid culture, the color is readily visible to naked eye in less than 24 hours of incubation. The DNA was codon-optimized for expression in E.coli and synthesized by the Korean company Bioneer Corporation.
Important: This part is not available in the registry yet, however, the same part is available from the registry with the RBS B0032: BBa_K1073023.
Usage and Biology
This part is useful as a reporter.
Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989049", https://parts.igem.org/Part:BBa_K1989050" or https://parts.igem.org/Part:BBa_K1989051".
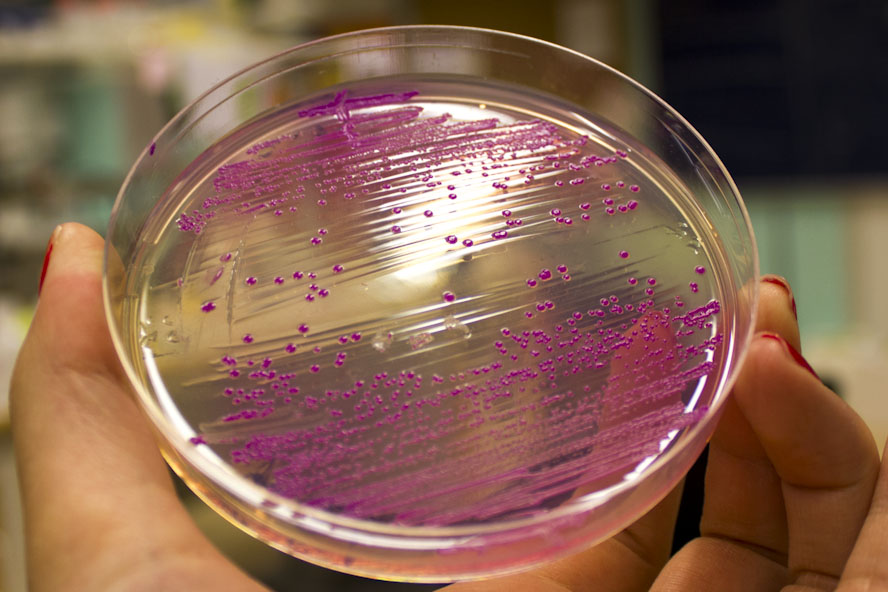
 iGEM12_Uppsala_University: Left picture: Visual color expression of eforRed (BBa_K592012).
iGEM12_Uppsala_University: Left picture: Visual color expression of eforRed (BBa_K592012).
Right picture: The Uppsala chromoprotein collection and RFP. The image shows pellets of E coli expressing chromoproteins eforRed BBa_K592012, RFP BBa_E1010, cjBlue BBa_K592011, aeBlue BBa_K864401, amilGFP BBa_K592010 and amilCP BBa_K592009.
iGEM17_SJTU_BioX_Shanghai:
J23119+Target3+eforRed BBa_K2285012
J23119+Target1+eforRed BBa_K2285016
An improved part has been constructed. Since this part is a coding sequence, we added a RBS which is on the upstream of sfGFP BBa_K515005 and terminator BBa_B1006 by PCR. What's more, our device have a stem-loop structure(called Target) following the constitutive promoter J23119, to achieve further control of eforRed expression.
References
[http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, N. O., et al. 2008. Diversity and evolution of coral fluorescent proteins. PLoS One 3:e2680.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
